Modulation of allosteric behavior through adjustment of the differential stability of the two interacting domains in E. coli cAMP receptor protein
- PMID: 21782316
- PMCID: PMC3166978
- DOI: 10.1016/j.bpc.2011.06.015
Modulation of allosteric behavior through adjustment of the differential stability of the two interacting domains in E. coli cAMP receptor protein
Abstract
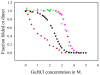
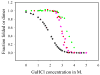
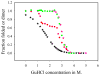
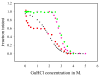
The communication mechanism(s) responsible for the allosteric behavior of E.coli cAMP binding receptor protein, CRP, is still a subject of intense investigation. As a tool to explore the communication mechanism, the mutations at various positions in the cAMP-binding (K52N, D53H, S62F and T127L) or the DNA- binding (H159L) domain or both (K52N/H159L) were generated. The sites and specific nature of side chain substitutions were defined by earlier genetic studies, the results of which show that these mutants have a similar phenotype i.e. they are activated without exogenous cAMP. Presently, no significant changes in the structures of WT and mutant CRPs have been observed. Hence, the pressing issue is to identify a physical parameter that reflects the effects of mutations. In this study, the stability of these various CRP species in the presence of GuHCl was monitored by three spectroscopic techniques, namely, CD, tryptophan fluorescence and FT-IR which could provide data on the stability of α-helices and β-strands separately. Results of this study led to the following conclusions: 1. The α-helices can be grouped into two families with different stabilities. Mutations exert a differential effect on the stability of helices as demonstrated by a biphasic unfolding curve for the helices. 2. Regardless of the locations of mutations, the effects can be communicated to the other domain resulting in a perturbation of the stability of both domains, although the effects are more significantly expressed in the stability of the helices. 3. Although in an earlier study [Gekko, et al. Biochemistry 43 (2004) 3844] we showed that cooperativity of cAMP binding is generally correlated to the global dynamics of the protein and DNA binding affinity, in this study we found that generally there is no clear correlation between functional energetics and stability of secondary structures. Thus, results of this study imply that modulation of allostery in CRP is entropic in nature.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures


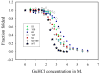
 , D53H;
, D53H;
 , H159L;
, H159L;
 , S62F; □, T127L;
, S62F; □, T127L;
 , K52N; ■, K52N/H159L; ★, WT.
, K52N; ■, K52N/H159L; ★, WT.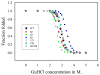
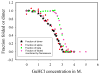
 , fraction of folded α-helix as monitored by FT-IR;
, fraction of folded α-helix as monitored by FT-IR;
 , fraction of folded β-strand as monitored by FT-IR;
, fraction of folded β-strand as monitored by FT-IR;
 , fraction of folded CRP as monitored by fluorescence.
, fraction of folded CRP as monitored by fluorescence.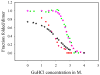

Similar articles
-
Communications between the high-affinity cyclic nucleotide binding sites in E. coli cyclic AMP receptor protein: effect of single site mutations.Biochemistry. 2002 Oct 1;41(39):11857-67. doi: 10.1021/bi026099z. Biochemistry. 2002. PMID: 12269830
-
A linear correlation between the energetics of allosteric communication and protein flexibility in the Escherichia coli cyclic AMP receptor protein revealed by mutation-induced changes in compressibility and amide hydrogen-deuterium exchange.Biochemistry. 2004 Apr 6;43(13):3844-52. doi: 10.1021/bi036271e. Biochemistry. 2004. PMID: 15049691
-
Interplay between site-specific mutations and cyclic nucleotides in modulating DNA recognition by Escherichia coli cyclic AMP receptor protein.Biochemistry. 2004 Jul 20;43(28):8901-10. doi: 10.1021/bi0499359. Biochemistry. 2004. PMID: 15248748
-
Syn, anti, and finally both conformations of cyclic AMP are involved in the CRP-dependent transcription initiation mechanism in E. coli lac operon.Cell Biochem Funct. 2008 Jun;26(4):399-405. doi: 10.1002/cbf.1462. Cell Biochem Funct. 2008. PMID: 18338329 Review.
-
Role of allosteric changes in cyclic AMP receptor protein function.Subcell Biochem. 1995;24:303-21. doi: 10.1007/978-1-4899-1727-0_10. Subcell Biochem. 1995. PMID: 7900180 Review. No abstract available.
Cited by
-
Asymmetric configurations in a reengineered homodimer reveal multiple subunit communication pathways in protein allostery.J Biol Chem. 2017 Apr 14;292(15):6086-6093. doi: 10.1074/jbc.M117.776047. Epub 2017 Feb 10. J Biol Chem. 2017. PMID: 28188293 Free PMC article.
-
Structural and energetic basis of allostery.Annu Rev Biophys. 2012;41:585-609. doi: 10.1146/annurev-biophys-050511-102319. Annu Rev Biophys. 2012. PMID: 22577828 Free PMC article. Review.
-
What Mutagenesis Can and Cannot Reveal About Allostery.Biophys J. 2016 May 10;110(9):1912-23. doi: 10.1016/j.bpj.2016.03.021. Biophys J. 2016. PMID: 27166800 Free PMC article. Review.
-
Identifying Allosteric Hotspots in Mycobacterium tuberculosis cAMP Receptor Protein through Structural Homology.Biochemistry. 2025 Feb 18;64(4):801-811. doi: 10.1021/acs.biochem.4c00723. Epub 2025 Jan 31. Biochemistry. 2025. PMID: 39887300 Free PMC article.
References
-
- Adhya S, Garges S. Positive control. J Biol Chem. 1990;265:10797–10800. - PubMed
-
- Busby S, Buc H. Positive regulation of gene expression by cyclic AMP and its receptor protein in Escherichia coli. Microb Sci. 1987;4:371–375. - PubMed
-
- de Crombrugghe B, Busby S, Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984;224:831–838. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

