The power of correlative microscopy: multi-modal, multi-scale, multi-dimensional
- PMID: 21782417
- PMCID: PMC3189301
- DOI: 10.1016/j.sbi.2011.06.010
The power of correlative microscopy: multi-modal, multi-scale, multi-dimensional
Abstract
Correlative microscopy is a sophisticated approach that combines the capabilities of typically separate, but powerful microscopy platforms: often including, but not limited, to conventional light, confocal and super-resolution microscopy, atomic force microscopy, transmission and scanning electron microscopy, magnetic resonance imaging and micro/nano CT (computed tomography). When targeting rare or specific events within large populations or tissues, correlative microscopy is increasingly being recognized as the method of choice. Furthermore, this multi-modal assimilation of technologies provides complementary and often unique information, such as internal and external spatial, structural, biochemical and biophysical details from the same targeted sample. The development of a continuous stream of cutting-edge applications, probes, preparation methodologies, hardware and software developments will enable realization of the full potential of correlative microscopy.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures


References
-
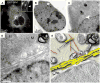
- Brown E, Verkade P. The use of markers for correlative light electron microscopy. Protoplasma. 2010;244:91–97. - PubMed
-
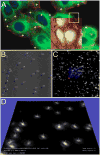
- Mironov AA, Beznoussenko GV. Correlative microscopy: a potent tool for the study of rare or unique cellular and tissue events. Journal of microscopy. 2009;235:308–321. - PubMed
-
- Brown E, Mantell J, Carter D, Tilly G, Verkade P. Studying intracellular transport using high-pressure freezing and Correlative Light Electron Microscopy. Seminars in cell & developmental biology. 2009;20:910–919. With HeLa cells and combining live-cell time-lapse microscopy and high-pressure freezing to follow endocytosis, the authors showed merits of STEM over TEM for visualization of individual quantun dot and gold conjugated probes. - PubMed
-
- Muller-Reichert T, Mantler J, Srayko M, O'Toole E. Electron microscopy of the early Caenorhabditis elegans embryo. Journal of microscopy. 2008;230:297–307. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

