Diversity and plasticity of the intracellular plant pathogen and insect symbiont "Candidatus Liberibacter asiaticus" as revealed by hypervariable prophage genes with intragenic tandem repeats
- PMID: 21784907
- PMCID: PMC3187138
- DOI: 10.1128/AEM.05111-11
Diversity and plasticity of the intracellular plant pathogen and insect symbiont "Candidatus Liberibacter asiaticus" as revealed by hypervariable prophage genes with intragenic tandem repeats
Abstract
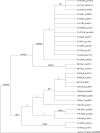
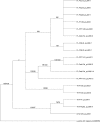
"Candidatus Liberibacter asiaticus" is a psyllid-transmitted, phloem-limited alphaproteobacterium and the most prevalent species of "Ca. Liberibacter" associated with a devastating worldwide citrus disease known as huanglongbing (HLB). Two related and hypervariable genes (hyv(I) and hyv(II)) were identified in the prophage regions of the Psy62 "Ca. Liberibacter asiaticus" genome. Sequence analyses of the hyv(I) and hyv(II) genes in 35 "Ca. Liberibacter asiaticus" DNA isolates collected globally revealed that the hyv(I) gene contains up to 12 nearly identical tandem repeats (NITRs, 132 bp) and 4 partial repeats, while hyv(II) contains up to 2 NITRs and 4 partial repeats and shares homology with hyv(I). Frequent deletions or insertions of these repeats within the hyv(I) and hyv(II) genes were observed, none of which disrupted the open reading frames. Sequence conservation within the individual repeats but an extensive variation in repeat numbers, rearrangement, and the sequences flanking the repeat region indicate the diversity and plasticity of "Ca. Liberibacter asiaticus" bacterial populations in the world. These differences were found not only in samples of distinct geographical origins but also in samples from a single origin and even from a single "Ca. Liberibacter asiaticus"-infected sample. This is the first evidence of different "Ca. Liberibacter asiaticus" populations coexisting in a single HLB-affected sample. The Florida "Ca. Liberibacter asiaticus" isolates contain both hyv(I) and hyv(II), while all other global "Ca. Liberibacter asiaticus" isolates contain either one or the other. Interclade assignments of the putative Hyv(I) and Hyv(II) proteins from Florida isolates with other global isolates in phylogenetic trees imply multiple "Ca. Liberibacter asiaticus" populations in the world and a multisource introduction of the "Ca. Liberibacter asiaticus" bacterium into Florida.
Figures


References
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

