Persistent, toxin-antitoxin system-independent, tetracycline resistance-encoding plasmid from a dairy Enterococcus faecium isolate
- PMID: 21784909
- PMCID: PMC3194845
- DOI: 10.1128/AEM.05168-11
Persistent, toxin-antitoxin system-independent, tetracycline resistance-encoding plasmid from a dairy Enterococcus faecium isolate
Abstract
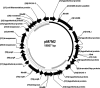
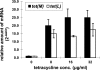
A tetracycline-resistant (Tet(r)) dairy Enterococcus faecium isolate designated M7M2 was found to carry both tet(M) and tet(L) genes on a 19.6-kb plasmid. After consecutive transfer in the absence of tetracycline, the resistance-encoding plasmid persisted in 99% of the progenies. DNA sequence analysis revealed that the 19.6-kb plasmid contained 28 open reading frames (ORFs), including a tet(M)-tet(L)-mob gene cluster, as well as a 10.6-kb backbone highly homologous (99.9%) to the reported plasmid pRE25, but without an identified toxin-antitoxin (TA) plasmid stabilization system. The derived backbone plasmid without the Tet(r) determinants exhibited a 100% retention rate in the presence of acridine orange, suggesting the presence of a TA-independent plasmid stabilization mechanism, with its impact on the persistence of a broad spectrum of resistance-encoding traits still to be elucidated. The tet(M)-tet(L) gene cluster from M7M2 was functional and transmissible and led to acquired resistance in Enterococcus faecalis OG1RF by electroporation and in Streptococcus mutans UA159 by natural transformation. Southern hybridization showed that both the tet(M) and tet(L) genes were integrated into the chromosome of S. mutans UA159, while the whole plasmid was transferred to and retained in E. faecalis OG1RF. Quantitative real-time reverse transcription-PCR (RT-PCR) indicated tetracycline-induced transcription of both the tet(M) and tet(L) genes of pM7M2. The results indicated that multiple mechanisms might have contributed to the persistence of antibiotic resistance-encoding genes and that the plasmids pM7M2, pIP816, and pRE25 are likely correlated evolutionarily.
Figures
References
-
- Allen H. K., et al. 2010. Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8:251–259 - PubMed
-
- Ausubel F. M., et al. 1989. Current protocols in molecular biology, vol. 1 Wiley Press, New York, NY
-
- Cocconcelli P. S., Cattivelli D., Gazzola S. 2003. Gene transfer of vancomycin and tetracycline resistances among Enterococcus faecalis during cheese and sausage fermentations. Int. J. Food Microbiol. 88:315–323 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

