Characterization of Escherichia coli UmuC active-site loops identifies variants that confer UV hypersensitivity
- PMID: 21784925
- PMCID: PMC3187380
- DOI: 10.1128/JB.05301-11
Characterization of Escherichia coli UmuC active-site loops identifies variants that confer UV hypersensitivity
Abstract
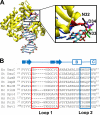
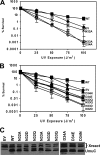
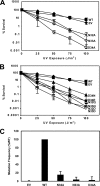
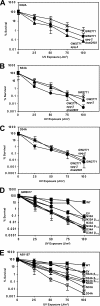
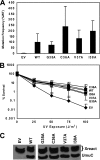
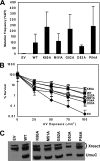
DNA is constantly exposed to chemical and environmental mutagens, causing lesions that can stall replication. In order to deal with DNA damage and other stresses, Escherichia coli utilizes the SOS response, which regulates the expression of at least 57 genes, including umuDC. The gene products of umuDC, UmuC and the cleaved form of UmuD, UmuD', form the specialized E. coli Y-family DNA polymerase UmuD'2C, or polymerase V (Pol V). Y-family DNA polymerases are characterized by their specialized ability to copy damaged DNA in a process known as translesion synthesis (TLS) and by their low fidelity on undamaged DNA templates. Y-family polymerases exhibit various specificities for different types of DNA damage. Pol V carries out TLS to bypass abasic sites and thymine-thymine dimers resulting from UV radiation. Using alanine-scanning mutagenesis, we probed the roles of two active-site loops composed of residues 31 to 38 and 50 to 54 in Pol V activity by assaying the function of single-alanine variants in UV-induced mutagenesis and for their ability to confer resistance to UV radiation. We find that mutations of the N-terminal residues of loop 1, N32, N33, and D34, confer hypersensitivity to UV radiation and to 4-nitroquinoline-N-oxide and significantly reduce Pol V-dependent UV-induced mutagenesis. Furthermore, mutating residues 32, 33, or 34 diminishes Pol V-dependent inhibition of recombination, suggesting that these mutations may disrupt an interaction of UmuC with RecA, which could also contribute to the UV hypersensitivity of cells expressing these variants.
Figures

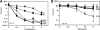
Similar articles
-
Point mutations in Escherichia coli DNA pol V that confer resistance to non-cognate DNA damage also alter protein-protein interactions.Mutat Res. 2015 Oct;780:1-14. doi: 10.1016/j.mrfmmm.2015.07.003. Epub 2015 Jul 13. Mutat Res. 2015. PMID: 26218456
-
Characterization of novel alleles of the Escherichia coli umuDC genes identifies additional interaction sites of UmuC with the beta clamp.J Bacteriol. 2009 Oct;191(19):5910-20. doi: 10.1128/JB.00292-09. Epub 2009 Jul 24. J Bacteriol. 2009. PMID: 19633075 Free PMC article.
-
Regulation of Mutagenic DNA Polymerase V Activation in Space and Time.PLoS Genet. 2015 Aug 28;11(8):e1005482. doi: 10.1371/journal.pgen.1005482. eCollection 2015 Aug. PLoS Genet. 2015. PMID: 26317348 Free PMC article.
-
Escherichia coli Y family DNA polymerases.Front Biosci (Landmark Ed). 2011 Jun 1;16(8):3164-82. doi: 10.2741/3904. Front Biosci (Landmark Ed). 2011. PMID: 21622227 Review.
-
Mutations for Worse or Better: Low-Fidelity DNA Synthesis by SOS DNA Polymerase V Is a Tightly Regulated Double-Edged Sword.Biochemistry. 2016 Apr 26;55(16):2309-18. doi: 10.1021/acs.biochem.6b00117. Epub 2016 Apr 12. Biochemistry. 2016. PMID: 27043933 Free PMC article. Review.
Cited by
-
Multiple strategies for translesion synthesis in bacteria.Cells. 2012 Oct 15;1(4):799-831. doi: 10.3390/cells1040799. Cells. 2012. PMID: 24710531 Free PMC article.
-
Altering the N-terminal arms of the polymerase manager protein UmuD modulates protein interactions.PLoS One. 2017 Mar 8;12(3):e0173388. doi: 10.1371/journal.pone.0173388. eCollection 2017. PLoS One. 2017. PMID: 28273172 Free PMC article.
-
A single residue unique to DinB-like proteins limits formation of the polymerase IV multiprotein complex in Escherichia coli.J Bacteriol. 2013 Mar;195(6):1179-93. doi: 10.1128/JB.01349-12. Epub 2013 Jan 4. J Bacteriol. 2013. PMID: 23292773 Free PMC article.
-
Structural model of the Y-Family DNA polymerase V/RecA mutasome.J Mol Graph Model. 2013 Feb;39:133-44. doi: 10.1016/j.jmgm.2012.09.005. Epub 2012 Nov 27. J Mol Graph Model. 2013. PMID: 23266508 Free PMC article.
-
Contribution of increased mutagenesis to the evolution of pollutants-degrading indigenous bacteria.PLoS One. 2017 Aug 4;12(8):e0182484. doi: 10.1371/journal.pone.0182484. eCollection 2017. PLoS One. 2017. PMID: 28777807 Free PMC article.
References
-
- Beard W. A., Wilson S. H. 2003. Structural insights into the origins of DNA polymerase fidelity. Structure 11:489–496 - PubMed
-
- Becherel O. J., Fuchs R. P. 1999. SOS mutagenesis results from up-regulation of translesion synthesis. J. Mol. Biol. 294:299–306 - PubMed
-
- Becherel O. J., Fuchs R. P. P., Wagner J. 2002. Pivotal role of the beta-clamp in translesion DNA synthesis and mutagenesis in E. coli cells. DNA Repair (Amst.) 1:703–708 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

