Quorum sensing contributes to natural transformation of Vibrio cholerae in a species-specific manner
- PMID: 21784943
- PMCID: PMC3165701
- DOI: 10.1128/JB.05396-11
Quorum sensing contributes to natural transformation of Vibrio cholerae in a species-specific manner
Abstract
Although it is a human pathogen, Vibrio cholerae is a regular member of aquatic habitats, such as coastal regions and estuaries. Within these environments, V. cholerae often takes advantage of the abundance of zooplankton and their chitinous molts as a nutritious surface on which the bacteria can form biofilms. Chitin also induces the developmental program of natural competence for transformation in several species of the genus Vibrio. In this study, we show that V. cholerae does not distinguish between species-specific and non-species-specific DNA at the level of DNA uptake. This is in contrast to what has been shown for other Gram-negative bacteria, such as Neisseria gonorrhoeae and Haemophilus influenzae. However, species specificity with respect to natural transformation still occurs in V. cholerae. This is based on a positive correlation between quorum sensing and natural transformation. Using mutant-strain analysis, cross-feeding experiments, and synthetic cholera autoinducer-1 (CAI-1), we provide strong evidence that the species-specific signaling molecule CAI-1 plays a major role in natural competence for transformation. We suggest that CAI-1 can be considered a competence pheromone.
Copyright © 2011, American Society for Microbiology. All Rights Reserved.
Figures

 ) is comparable to this LCD state. Strain A1552ΔluxO (★) is similar to an HCD state because it lacks the posttranscriptional inhibition of HapR synthesis. Abbreviations: ∼, phosphorylated; de∼, dephosphorylated. This scheme is based on findings in references , , and .
) is comparable to this LCD state. Strain A1552ΔluxO (★) is similar to an HCD state because it lacks the posttranscriptional inhibition of HapR synthesis. Abbreviations: ∼, phosphorylated; de∼, dephosphorylated. This scheme is based on findings in references , , and .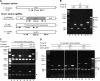
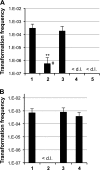
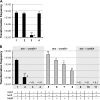
References
-
- Allemand J. F., Maier B. 2009. Bacterial translocation motors investigated by single molecule techniques. FEMS Microbiol. Rev. 33:593–610 - PubMed
-
- Berge M., Mortier-Barriere I., Martin B., Claverys J. P. 2003. Transformation of Streptococcus pneumoniae relies on DprA- and RecA-dependent protection of incoming DNA single strands. Mol. Microbiol. 50:527–536 - PubMed
-
- Bertuzzo E., et al. 2011. Prediction of the spatial evolution and effects of control measures for the unfolding Haiti cholera outbreak. Geophys. Res. Lett. 38:L06403.doi:10.1029/2011GL046823. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

