Enhancers regulate progression of development in mammalian cells
- PMID: 21785139
- PMCID: PMC3203619
- DOI: 10.1093/nar/gkr602
Enhancers regulate progression of development in mammalian cells
Abstract
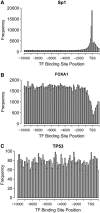
During development and differentiation of an organism, accurate gene regulation is central for cells to maintain and balance their differentiation processes. Transcriptional interactions between cis-acting DNA elements such as promoters and enhancers are the basis for precise and balanced transcriptional regulation. We identified modules of combinations of binding sites in proximal and distal regulatory regions upstream of all transcription start sites (TSSs) in silico and applied these modules to gene expression time-series of mouse embryonic development and differentiation of human stem cells. In addition to tissue-specific regulation controlled by combinations of transcription factors (TFs) binding at promoters, we observed that in particular the combination of TFs binding at promoters together with TFs binding at the respective enhancers regulate highly specifically temporal progression during development: whereas 40% of TFs were specific for time intervals, 79% of TF pairs and even 97% of promoter-enhancer modules showed specificity for single time intervals of the human stem cells. Predominantly SP1 and E2F contributed to temporal specificity at promoters and the forkhead (FOX) family of TFs at enhancer regions. Altogether, we characterized three classes of TFs: with binding sites being enriched at the TSS (like SP1), depleted at the TSS (like FOX), and rather uniformly distributed.
Figures



Similar articles
-
Probing transcription factor combinatorics in different promoter classes and in enhancers.BMC Genomics. 2019 Feb 1;20(1):103. doi: 10.1186/s12864-018-5408-0. BMC Genomics. 2019. PMID: 30709337 Free PMC article.
-
Inferring dynamic gene regulatory networks in cardiac differentiation through the integration of multi-dimensional data.BMC Bioinformatics. 2015 Mar 7;16:74. doi: 10.1186/s12859-015-0460-0. BMC Bioinformatics. 2015. PMID: 25887857 Free PMC article.
-
Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers.Genome Res. 2010 May;20(5):565-77. doi: 10.1101/gr.104471.109. Epub 2010 Apr 2. Genome Res. 2010. PMID: 20363979 Free PMC article.
-
Understanding fundamental principles of enhancer biology at a model locus: Analysing the structure and function of an enhancer cluster at the α-globin locus.Bioessays. 2023 Oct;45(10):e2300047. doi: 10.1002/bies.202300047. Epub 2023 Jul 5. Bioessays. 2023. PMID: 37404089 Free PMC article. Review.
-
Combinatorial function of transcription factors and cofactors.Curr Opin Genet Dev. 2017 Apr;43:73-81. doi: 10.1016/j.gde.2016.12.007. Epub 2017 Jan 19. Curr Opin Genet Dev. 2017. PMID: 28110180 Review.
Cited by
-
Evaluation of methylation status of the eNOS promoter at birth in relation to childhood bone mineral content.Calcif Tissue Int. 2012 Feb;90(2):120-7. doi: 10.1007/s00223-011-9554-5. Epub 2011 Dec 8. Calcif Tissue Int. 2012. PMID: 22159788 Free PMC article.
-
Characterizing protein interactions employing a genome-wide siRNA cellular phenotyping screen.PLoS Comput Biol. 2014 Sep 25;10(9):e1003814. doi: 10.1371/journal.pcbi.1003814. eCollection 2014 Sep. PLoS Comput Biol. 2014. PMID: 25255318 Free PMC article.
-
The genomic signature of trait-associated variants.BMC Genomics. 2013 Feb 18;14:108. doi: 10.1186/1471-2164-14-108. BMC Genomics. 2013. PMID: 23418889 Free PMC article.
-
Sp1 trans-activates and is required for maximal aldosterone induction of the αENaC gene in collecting duct cells.Am J Physiol Renal Physiol. 2013 Sep 1;305(5):F653-62. doi: 10.1152/ajprenal.00177.2013. Epub 2013 Jun 26. Am J Physiol Renal Physiol. 2013. PMID: 23804453 Free PMC article.
-
Childhood bone mineral content is associated with methylation status of the RXRA promoter at birth.J Bone Miner Res. 2014 Mar;29(3):600-7. doi: 10.1002/jbmr.2056. J Bone Miner Res. 2014. PMID: 23907847 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

