esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function
- PMID: 21785422
- PMCID: PMC3155811
- DOI: 10.1038/ncb2285
esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function
Abstract
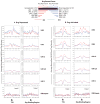
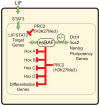
Signalling by the cytokine LIF and its downstream transcription factor, STAT3, prevents differentiation of pluripotent embryonic stem cells (ESCs). This contrasts with most cell types where STAT3 signalling induces differentiation. We find that STAT3 binding across the pluripotent genome is dependent on Brg1, the ATPase subunit of a specialized chromatin remodelling complex (esBAF) found in ESCs. Brg1 is required to establish chromatin accessibility at STAT3 binding targets, preparing these sites to respond to LIF signalling. Brg1 deletion leads to rapid polycomb (PcG) binding and H3K27me3-mediated silencing of many Brg1-activated targets genome wide, including the target genes of the LIF signalling pathway. Hence, one crucial role of Brg1 in ESCs involves its ability to potentiate LIF signalling by opposing PcG. Contrary to expectations, Brg1 also facilitates PcG function at classical PcG targets, including all four Hox loci, reinforcing their repression in ESCs. Therefore, esBAF does not simply antagonize PcG. Rather, the two chromatin regulators act both antagonistically and synergistically with the common goal of supporting pluripotency.
Figures






Comment in
-
esBAF safeguards Stat3 binding to maintain pluripotency.Nat Cell Biol. 2011 Aug 1;13(8):886-8. doi: 10.1038/ncb2311. Nat Cell Biol. 2011. PMID: 21808242
References
-
- Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

