A refined molecular taxonomy of breast cancer
- PMID: 21785460
- PMCID: PMC3307061
- DOI: 10.1038/onc.2011.301
A refined molecular taxonomy of breast cancer
Abstract
The current histoclinical breast cancer classification is simple but imprecise. Several molecular classifications of breast cancers based on expression profiling have been proposed as alternatives. However, their reliability and clinical utility have been repeatedly questioned, notably because most of them were derived from relatively small initial patient populations. We analyzed the transcriptomes of 537 breast tumors using three unsupervised classification methods. A core subset of 355 tumors was assigned to six clusters by all three methods. These six subgroups overlapped with previously defined molecular classes of breast cancer, but also showed important differences, notably the absence of an ERBB2 subgroup and the division of the large luminal ER+ group into four subgroups, two of them being highly proliferative. Of the six subgroups, four were ER+/PR+/AR+, one was ER-/PR-/AR+ and one was triple negative (AR-/ER-/PR-). ERBB2-amplified tumors were split between the ER-/PR-/AR+ subgroup and the highly proliferative ER+ LumC subgroup. Importantly, each of these six molecular subgroups showed specific copy-number alterations. Gene expression changes were correlated to specific signaling pathways. Each of these six subgroups showed very significant differences in tumor grade, metastatic sites, relapse-free survival or response to chemotherapy. All these findings were validated on large external datasets including more than 3000 tumors. Our data thus indicate that these six molecular subgroups represent well-defined clinico-biological entities of breast cancer. Their identification should facilitate the detection of novel prognostic factors or therapeutical targets in breast cancer.
Figures
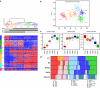

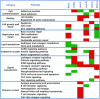

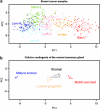
References
-
- Bertucci F, Finetti P, Cervera N, Maraninchi D, Viens P. Gene expression profiling and clinical outcome in breast cancer. Omics. 2006;10:429–443. - PubMed
-
- Bonnefoi H, Potti A, Delorenzi M, Mauriac L, Campone M, Tubiana-Hulin M, et al. Validation of gene signatures that predict the response of breast cancer to neoadjuvant chemotherapy: a substudy of the EORTC 10994/BIG 00-01 clinical trial. Lancet Oncol. 2007;8:1071–1078. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

