Absence of Runx3 expression in normal gastrointestinal epithelium calls into question its tumour suppressor function
- PMID: 21786422
- PMCID: PMC3258485
- DOI: 10.1002/emmm.201100168
Absence of Runx3 expression in normal gastrointestinal epithelium calls into question its tumour suppressor function
Abstract
The Runx3 transcription factor regulates cell fate decisions during embryonic development and in adults. It was previously reported that Runx3 is strongly expressed in embryonic and adult gastrointestinal tract (GIT) epithelium (Ep) and that its loss causes gastric cancer. More than 280 publications have based their research on these findings and concluded that Runx3 is indeed a tumour suppressor (TS). In stark contrast, using various measures, we found that Runx3 expression is undetectable in GIT Ep. Employing a variety of biochemical and genetic techniques, including analysis of Runx3-GFP and R26LacZ/Runx3(Cre) or R26tdTomato/Runx3(Cre) reporter strains, we readily detected Runx3 in GIT-embedded leukocytes, dorsal root ganglia, skeletal elements and hair follicles. However, none of these approaches revealed detectable Runx3 levels in GIT Ep. Moreover, our analysis of the original Runx3(LacZ/LacZ) mice used in the previously reported study failed to reproduce the GIT expression of Runx3. The lack of evidence for Runx3 expression in normal GIT Ep creates a serious challenge to the published data and undermines the notion that Runx3 is a TS involved in cancer pathogenesis.
Copyright © 2011 EMBO Molecular Medicine.
Figures

A. Scheme depicting the Runx3 gene and the targeted alleles used to generate the Kyoto- and Rehovot- Runx3LacZ/LacZ mice. The two promoters (P1 and P2) and the corresponding initiator ATGs are indicated. In Kyoto-Runx3LacZ/LacZ mice, the LacZ was inserted in frame into exon 4 of the gene creating a Runx3-LacZ fusion protein (Li et al, 2002). In Rehovot- Runx3LacZ/LacZ mice, the gene was disrupted by inserting an IRES-LacZ into exon 2 (Levanon et al, 2002).
B,C. The pattern of Runx3 expression at E14.5 revealed by whole mount LacZ staining of Rehovot-Runx3LacZ/+ embryo (B) and by IHC of WT embryo using a sagittal section reacted with Poly-G Ab (C). Runx3 is strongly expressed in whiskers (W), cartilage (C), thymus (T), DRG (D) and haematopoietic cells in the liver (L), but not in the GIT (G).
D,E. Isolated LacZ-stained GIT of E14.5 Runx3LacZ/+ embryo (D) and a transverse section of the intestine of a WT embryo (E) immunostained with poly-G Abs showing lack of detectable Runx3 in the epithelium (Ep).

Scheme showing Runx3 protein structure indicating the position of peptides used for generation of the five anti-Runx3 Abs (see Materials and Methods Section for details). Poly-G, Poly-SA-, Pep-J and GS are rabbit polyclonal Abs. Mono-G is a monoclonal Ab.
Transverse sections of adult WT (left) and Runx3−/− (right) small intestine immunostained with the Poly-G Ab. Runx3 was detected in WT GIT-embedded leukocytes (left) but not in Runx3−/− cells. No Runx3 was detected in WT GIT Ep.
Transverse sections of the small intestine of an adult CX3CR1GFP/+ mouse (Jung et al, 2000) double-stained with Poly-G and anti-GFP Abs. In adult GIT, Runx3 is expressed in dendritic cells (DCs) and intraepithelial leukocytes (IEL). CX3CR1GFP marks the GIT DCs. Runx3 was detected in GIT IEL (black nuclear staining, see arrows in the enlarged right panel). Double stained Runx3/GFP positive (brown cell membrane staining) depicts Runx3 expressing DCs. No Runx3 was detected in GIT Ep.
Three distinct anti-Runx3 Abs (From left to right: Poly-G, Poly-SA and Pep-J) detected Runx3 in small intestine Peyer's patch (PP) and leukocytes of adult mice, but not in GIT Ep.
Adult WT small intestine immunostained with Mono-G anti-Runx3 Ab (left panel) and with 4′-6-diamidino-2-phenylindole (DAPI) which stains the nuclei of both leukocytes and epithelial cells (middle panel) and a merged image of the two frames (right panel) revealing Runx3 expression in GIT-embeded IEL, but not in the Ep.
In E14.5 WT embryos Mono-G Abs detected Runx3 in DRG (left) but not in GIT (right). More details are presented in Fig S1 of Supporting information and in Supporting information text: evaluation of anti Runx3 Abs).
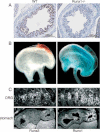
Sections of E16.5 forestomach of WT and Runx1-P2Neo/Neo embryos immunostained with anti-Runx1 Ab show Runx1 expression in WT but not in the negative control Runx1-P2Neo/Neo forestomach (adapted from Pozner et al, 2007).
Whole mount LacZ stained stomach of WT (left) and Runx1LacZ/+ (right) E14.5 embryos. LacZ staining was more prominent in the forestomach.
35S-RNA in situ hybridization analysis of Runx1 and Runx3 expression in embryonic DRG and stomach. DRG and stomach of E16.5 WT embryos were hybridized with a Runx3 probe (left panels) and a Runx1 probe (right panels). Both probes detected expression in the DRG (Runx1 in TrkA neurons and Runx3 in TrkC neurons), but only the Runx1 probe detected expression in gastric Ep. F, forestomach, G, glandular stomach (adapted from Brenner et al, 2004).
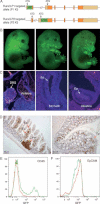
A. A scheme of the P1-AFP and P2-EGFP targeted alleles used to generate the Runx3-GFP reporter mice.
B. Whole mount view of Runx3P1-AFP/+, Runx3P1-AFP/P2-GFP and Runx3P2-GFP/+ (from left to right) E14.5 embryos.
C. Sagittal sections of DRG and GIT of E14.5 Runx3P1-AFP/P2-GFP embryos immunostained with anti-GFP Abs. GFP was detected in DRG and vertebrae, but not in gastric and intestinal epithelium (Ep).
D. Sections of adult Runx3P1-AFP/P2-GFP GIT (small intestine, left; colon, right) immunostained with anti-GFP Abs show GFP positive leukocytes in Peyer's patch (PP) and IEL and in leukocytes within the lamina propria, while the adjacent Ep is unstained.
E,F. Flow cytometric analysis of GFP expression in single cell-suspensions of GIT Ep of adult Runx3P1-AFP/P2-GFP mice. Histograms demonstrating EGFP/AFP expression in CD45+ IEL (E) or EpCAM+ epithelial cells (F) of Runx3P1-AFP/P2-GFP GIT (green) compared to WT (red). No GFP positive GIT epithelial cells (F) were detected. Results from one of four Runx3P1-AFP/P2-GFP and WT control mice with the same findings are shown. The relative high mean fluorescence intensity of both Runx3P1-AFP/P2-GFP and WT GIT epithelial cells was due to the known autofluorescence of epithelial cells (DaCosta et al, 2005).
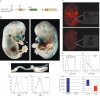
A. Scheme of the Runx3-GFP-Cre targeted allele used to create the Runx3Cre mice. The GFP-Cre cassette was inserted in frame into the SmaI site used for the generation of the Kyoto-Runx3LacZ/LacZ mice (see Fig 1A).
B,C. LacZ expression in whole mount E14.5 R26-LacZ/Runx3Cre embryo. The GIT, outlined in red (right panel), was excised, stretched and is shown magnified in (C). W, whiskers; C, cartilage; T, thymus; L, liver; HF, hair follicles.
D,E. Flow cytometric analysis of Runx3 expression (FDG serves as a fluorescence substrate for β-gal) in thymocytes and GIT epithelial cells of E16.5 R26-LacZ/Runx3Cre embryos. (D) Histograms demonstrating detection of CD45+/FDG+ double positive thymocytes (green) of R26-LacZ/Runx3Cre embryo compared to WT (red). (E) Absence of FDG+ cells in GIT epithelial cells of either WT (red) or R26-LacZ/Runx3Cre embryos (green). Results from one of four R26-LacZ/Runx3Cre and WT control embryos with same findings are shown.
F. tdTomato expression in whole mount E14.5 R26-tdTomato/Runx3Cre (upper panel) or R26-tdTomato (lower panel) embryos. Red tdTomato fluorescence is seen in cartilage of skeletal elements, whiskers and hair follicles. The GIT of both R26-tdTomato/Runx3Cre and R26-tdTomato embryos was excised, stretched and is shown magnified on the right.
G. Flow cytometric analysis of Runx3 expression (via tdTomato fluorescence) in thymocytes and GIT epithelial cells of E16.5 R26-tdTomato/Runx3Cre embryos.
H. Flow cytometric analysis of tdTomato in single cell-suspensions of GIT epithelium (Ep) of adult R26-tdTomato/Runx3Cre mice. Results from one of four adult R26-tdTomato/Runx3Cre mice with same findings are shown.
I. RT-qPCR analysis of Runx3 expression in splenic NK and GIT epithelial cells of adult WT mice. cDNAs of splenic CD45+ NK cells and of sorted EpCAM+CD45− GIT epithelial cells were analysed with Runx3, PBGD and HPRT TaqMan assays as detailed under Materials and Methods Section. Results were normalized to endogenous control genes and calculated relative to a calibrator. Left panel. Expression of Runx3 in CD45+NK cells is 2257 fold higher relative to the EpCAM+ epithelial cells as calibrator (p = 0.001). The right panel depicts the expression of Runx3 in CD45+NK cells and EpCAM+CD45− epithelial cells calculated relative to the PBGD as calibrator (p = 0.001). Of note, signals produced by the EpCAM+CD45− GIT epithelial sample were consistently detected after more than 35 cycles, 5 cycles higher than any other assay for an equivalent RNA input. By these standard criteria, we interpret the recorded values to mean there is effectively no true target for Runx3 in WT GIT Ep.

Comment in
-
RUNX3 is expressed in the epithelium of the gastrointestinal tract.EMBO Mol Med. 2012 Jul;4(7):541-2; author reply 543-4. doi: 10.1002/emmm.201100203. Epub 2012 May 31. EMBO Mol Med. 2012. PMID: 22648993 Free PMC article. No abstract available.
References
-
- Avraham KB, Levanon D, Negreanu V, Bernstein Y, Groner Y, Copeland NG, Jenkins NA. Mapping of the mouse homolog of the human runt domain gene, AML2, to the distal region of mouse chromosome 4. Genomics. 1995;25:603–605. - PubMed
-
- Bangsow C, Rubins N, Glusman G, Bernstein Y, Negreanu V, Goldenberg D, Lotem J, Ben-Asher E, Lancet D, Levanon D, et al. The RUNX3 gene—sequence, structure and regulated expression. Gene. 2001;279:221–232. - PubMed
-
- Bennecke M, Kriegl L, Bajbouj M, Retzlaff K, Robine S, Jung A, Arkan MC, Kirchner T, Greten FR. Ink4a/Arf and oncogene-induced senescence prevent tumor progression during alternative colorectal tumorigenesis. Cancer Cell. 2010;18:135–146. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

