Defining genome maintenance pathways using functional genomic approaches
- PMID: 21787120
- PMCID: PMC3144553
- DOI: 10.3109/10409238.2011.588938
Defining genome maintenance pathways using functional genomic approaches
Abstract
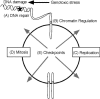
Genome maintenance activities including DNA repair, cell division cycle control, and checkpoint signaling pathways preserve genome integrity and prevent disease. Defects in these pathways cause birth defects, neurodegeneration, premature aging, and cancer. Recent technical advances in functional genomic approaches such as expression profiling, proteomics, and RNA interference (RNAi) technologies have rapidly expanded our knowledge of the proteins that work in these pathways. In this review, we examine the use of these high-throughput methodologies in higher eukaryotic organisms for the interrogation of genome maintenance activities.
Figures

Similar articles
-
Cell cycle checkpoints: the role and evaluation for early diagnosis of senescence, cardiovascular, cancer, and neurodegenerative diseases.Amino Acids. 2007;32(3):359-71. doi: 10.1007/s00726-006-0473-0. Epub 2006 Nov 30. Amino Acids. 2007. PMID: 17136506 Review.
-
Functional proteomics: mapping protein-protein interactions and pathways.Curr Opin Mol Ther. 2002 Jun;4(3):210-5. Curr Opin Mol Ther. 2002. PMID: 12139305 Review.
-
Chromatin and the genome integrity network.Nat Rev Genet. 2013 Jan;14(1):62-75. doi: 10.1038/nrg3345. Nat Rev Genet. 2013. PMID: 23247436 Free PMC article. Review.
-
Functional genomic screens identify CINP as a genome maintenance protein.Proc Natl Acad Sci U S A. 2009 Nov 17;106(46):19304-9. doi: 10.1073/pnas.0909345106. Epub 2009 Nov 4. Proc Natl Acad Sci U S A. 2009. PMID: 19889979 Free PMC article.
-
Chromatin relaxation-mediated induction of p19INK4d increases the ability of cells to repair damaged DNA.PLoS One. 2013 Apr 12;8(4):e61143. doi: 10.1371/journal.pone.0061143. Print 2013. PLoS One. 2013. PMID: 23593412 Free PMC article.
Cited by
-
DNA damage response: three levels of DNA repair regulation.Cold Spring Harb Perspect Biol. 2013 Aug 1;5(8):a012724. doi: 10.1101/cshperspect.a012724. Cold Spring Harb Perspect Biol. 2013. PMID: 23813586 Free PMC article. Review.
-
Genome maintenance and human longevity.Curr Opin Genet Dev. 2014 Jun;26:105-15. doi: 10.1016/j.gde.2014.07.002. Epub 2014 Aug 28. Curr Opin Genet Dev. 2014. PMID: 25151201 Free PMC article. Review.
-
Alterations induced by the PML-RARα oncogene revealed by image cross correlation spectroscopy.Biophys J. 2022 Nov 15;121(22):4358-4367. doi: 10.1016/j.bpj.2022.10.003. Epub 2022 Oct 4. Biophys J. 2022. PMID: 36196056 Free PMC article.
References
-
- Amundson SA, Bittner M, Fornace AJ., Jr. Functional genomics as a window on radiation stress signaling. Oncogene. 2003;22:5828–33. - PubMed
-
- Baryshnikova A, Costanzo M, Dixon S, Vizeacoumar FJ, Myers CL, Andrews B, Boone C. Synthetic genetic array (SGA) analysis in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Methods Enzymol. 2010;470:145–79. - PubMed
-
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–74. - PubMed
-
- Bennett CB, Lewis LK, Karthikeyan G, Lobachev KS, Jin YH, Sterling JF, Snipe JR, Resnick MA. Genes required for ionizing radiation resistance in yeast. Nat Genet. 2001;29:426–34. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
