Quantification of codon selection for comparative bacterial genomics
- PMID: 21787402
- PMCID: PMC3162537
- DOI: 10.1186/1471-2164-12-374
Quantification of codon selection for comparative bacterial genomics
Abstract
Background: Statistics measuring codon selection seek to compare genes by their sensitivity to selection for translational efficiency, but existing statistics lack a model for testing the significance of differences between genes. Here, we introduce a new statistic for measuring codon selection, the Adaptive Codon Enrichment (ACE).
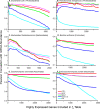
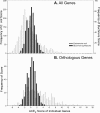
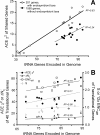
Results: This statistic represents codon usage bias in terms of a probabilistic distribution, quantifying the extent that preferred codons are over-represented in the gene of interest relative to the mean and variance that would result from stochastic sampling of codons. Expected codon frequencies are derived from the observed codon usage frequencies of a broad set of genes, such that they are likely to reflect nonselective, genome wide influences on codon usage (e.g. mutational biases). The relative adaptiveness of synonymous codons is deduced from the frequency of codon usage in a pre-selected set of genes relative to the expected frequency. The ACE can predict both transcript abundance during rapid growth and the rate of synonymous substitutions, with accuracy comparable to or greater than existing metrics. We further examine how the composition of reference gene sets affects the accuracy of the statistic, and suggest methods for selecting appropriate reference sets for any genome, including bacteriophages. Finally, we demonstrate that the ACE may naturally be extended to quantify the genome-wide influence of codon selection in a manner that is sensitive to a large fraction of codons in the genome. This reveals substantial variation among genomes, correlated with the tRNA gene number, even among groups of bacteria where previously proposed whole-genome measures show little variation.
Conclusions: The statistical framework of the ACE allows rigorous comparison of the level of codon selection acting on genes, both within a genome and between genomes.
Figures
Similar articles
-
Codon Usage Optimization in the Prokaryotic Tree of Life: How Synonymous Codons Are Differentially Selected in Sequence Domains with Different Expression Levels and Degrees of Conservation.mBio. 2020 Jul 21;11(4):e00766-20. doi: 10.1128/mBio.00766-20. mBio. 2020. PMID: 32694138 Free PMC article.
-
The Impact of Selection at the Amino Acid Level on the Usage of Synonymous Codons.G3 (Bethesda). 2017 Mar 10;7(3):967-981. doi: 10.1534/g3.116.038125. G3 (Bethesda). 2017. PMID: 28122952 Free PMC article.
-
Genome-wide codon usage profiling of ocular infective Chlamydia trachomatis serovars and drug target identification.J Biomol Struct Dyn. 2018 Jun;36(8):1979-2003. doi: 10.1080/07391102.2017.1343685. Epub 2017 Jul 4. J Biomol Struct Dyn. 2018. PMID: 28627970
-
Synonymous codon usage in bacteria.Curr Issues Mol Biol. 2001 Oct;3(4):91-7. Curr Issues Mol Biol. 2001. PMID: 11719972 Review.
-
The Yin and Yang of codon usage.Hum Mol Genet. 2016 Oct 1;25(R2):R77-R85. doi: 10.1093/hmg/ddw207. Epub 2016 Jun 27. Hum Mol Genet. 2016. PMID: 27354349 Free PMC article. Review.
Cited by
-
Metagenomic and phylogenetic analyses reveal gene-level selection constrained by bacterial phylogeny, surrounding oxalate metabolism in the gut microbiota.mSphere. 2025 Jun 25;10(6):e0091324. doi: 10.1128/msphere.00913-24. Epub 2025 May 13. mSphere. 2025. PMID: 40358144 Free PMC article.
-
Genomics and proteomics of mycobacteriophage patience, an accidental tourist in the Mycobacterium neighborhood.mBio. 2014 Dec 2;5(6):e02145. doi: 10.1128/mBio.02145-14. mBio. 2014. PMID: 25467442 Free PMC article.
-
The Code of Silence: Widespread Associations Between Synonymous Codon Biases and Gene Function.J Mol Evol. 2016 Jan;82(1):65-73. doi: 10.1007/s00239-015-9714-8. Epub 2015 Nov 4. J Mol Evol. 2016. PMID: 26538122 Review.
-
Inferring gene function from evolutionary change in signatures of translation efficiency.Genome Biol. 2014 Mar 3;15(3):R44. doi: 10.1186/gb-2014-15-3-r44. Genome Biol. 2014. PMID: 24580753 Free PMC article.
-
Ecological adaptation in bacteria: speciation driven by codon selection.Mol Biol Evol. 2012 Dec;29(12):3669-83. doi: 10.1093/molbev/mss171. Epub 2012 Jun 26. Mol Biol Evol. 2012. PMID: 22740635 Free PMC article.
References
-
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981;151:389–409. doi: 10.1016/0022-2836(81)90003-6. - DOI - PubMed
-
- Sharp PM, Li WH. The rate of synonymous substitution in enterobacterial genes is inversely related to codon usage bias. Mol Biol Evol. 1987;4:222–230. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

