Campylobacter jejuni dsb gene expression is regulated by iron in a Fur-dependent manner and by a translational coupling mechanism
- PMID: 21787430
- PMCID: PMC3167755
- DOI: 10.1186/1471-2180-11-166
Campylobacter jejuni dsb gene expression is regulated by iron in a Fur-dependent manner and by a translational coupling mechanism
Erratum in
- BMC Microbiol. 2012;12:58
Abstract
Background: Many bacterial extracytoplasmic proteins are stabilized by intramolecular disulfide bridges that are formed post-translationally between their cysteine residues. This protein modification plays an important role in bacterial pathogenesis, and is facilitated by the Dsb (disulfide bond) family of the redox proteins. These proteins function in two parallel pathways in the periplasmic space: an oxidation pathway and an isomerization pathway. The Dsb oxidative pathway in Campylobacter jejuni is more complex than the one in the laboratory E. coli K-12 strain.
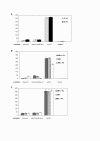
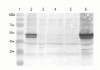
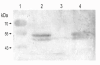
Results: In the C. jejuni 81-176 genome, the dsb genes of the oxidative pathway are arranged in three transcriptional units: dsbA2-dsbB-astA, dsbA1 and dba-dsbI. Their transcription responds to an environmental stimulus - iron availability - and is regulated in a Fur-dependent manner. Fur involvement in dsb gene regulation was proven by a reporter gene study in a C. jejuni wild type strain and its isogenic fur mutant. An electrophoretic mobility shift assay (EMSA) confirmed that analyzed genes are members of the Fur regulon but each of them is regulated by a disparate mechanism, and both the iron-free and the iron-complexed Fur are able to bind in vitro to the C. jejuni promoter regions. This study led to identification of a new iron- and Fur-regulated promoter that drives dsbA1 gene expression in an indirect way. Moreover, the present work documents that synthesis of DsbI oxidoreductase is controlled by the mechanism of translational coupling. The importance of a secondary dba-dsbI mRNA structure for dsbI mRNA translation was verified by estimating individual dsbI gene expression from its own promoter.
Conclusions: The present work shows that iron concentration is a significant factor in dsb gene transcription. These results support the concept that iron concentration - also through its influence on dsb gene expression - might control the abundance of extracytoplasmic proteins during different stages of infection. Our work further shows that synthesis of the DsbI membrane oxidoreductase is controlled by a translational coupling mechanism. The dba expression is not only essential for the translation of the downstream dsbI gene, but also Dba protein that is produced might regulate the activity and/or stability of DsbI.
Figures



References
-
- Woodall CA, Jones MA, Barrow PA, Hinds J, Marsden GL, Kelly DJ, Dorrell N, Wren BW, Maskell DJ. Campylobacter jejuni gene expression in the chick cecum: evidence for adaptation to a low-oxygen environment. Infect Immun. 2005;73(8):5278–5285. doi: 10.1128/IAI.73.8.5278-5285.2005. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

