Acetylation of H2A.Z is a key epigenetic modification associated with gene deregulation and epigenetic remodeling in cancer
- PMID: 21788347
- PMCID: PMC3266038
- DOI: 10.1101/gr.118919.110
Acetylation of H2A.Z is a key epigenetic modification associated with gene deregulation and epigenetic remodeling in cancer
Abstract
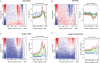
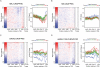
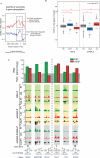
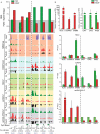
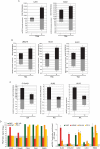
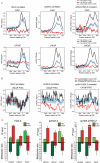
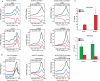
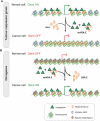
Histone H2A.Z (H2A.Z) is an evolutionarily conserved H2A variant implicated in the regulation of gene expression; however, its role in transcriptional deregulation in cancer remains poorly understood. Using genome-wide studies, we investigated the role of promoter-associated H2A.Z and acetylated H2A.Z (acH2A.Z) in gene deregulation and its relationship with DNA methylation and H3K27me3 in prostate cancer. Our results reconcile the conflicting reports of positive and negative roles for histone H2A.Z and gene expression states. We find that H2A.Z is enriched in a bimodal distribution at nucleosomes, surrounding the transcription start sites (TSSs) of both active and poised gene promoters. In addition, H2A.Z spreads across the entire promoter of inactive genes in a deacetylated state. In contrast, acH2A.Z is only localized at the TSSs of active genes. Gene deregulation in cancer is also associated with a reorganization of acH2A.Z and H2A.Z nucleosome occupancy across the promoter region and TSS of genes. Notably, in cancer cells we find that a gain of acH2A.Z at the TSS occurs with an overall decrease of H2A.Z levels, in concert with oncogene activation. Furthermore, deacetylation of H2A.Z at TSSs is increased with silencing of tumor suppressor genes. We also demonstrate that acH2A.Z anti-correlates with promoter H3K27me3 and DNA methylation. We show for the first time, that acetylation of H2A.Z is a key modification associated with gene activity in normal cells and epigenetic gene deregulation in tumorigenesis.
Figures
References
-
- Balasubramanyam K, Swaminathan V, Ranganathan A, Kundu TK 2003. Small molecule modulators of histone acetyltransferase p300. J Biol Chem 278: 19134–19140 - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 - PubMed
-
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ 3rd, Gingeras TR, et al. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120: 169–181 - PubMed
-
- Campos EI, Reinberg D 2009. Histones: annotating chromatin. Annu Rev Genet 43: 559–599 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
