MicroRNA regulation of molecular networks mapped by global microRNA, mRNA, and protein expression in activated T lymphocytes
- PMID: 21788445
- PMCID: PMC3159804
- DOI: 10.4049/jimmunol.1101233
MicroRNA regulation of molecular networks mapped by global microRNA, mRNA, and protein expression in activated T lymphocytes
Abstract
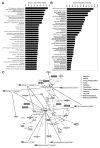
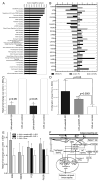
MicroRNAs (miRNAs) regulate specific immune mechanisms, but their genome-wide regulation of T lymphocyte activation is largely unknown. We performed a multidimensional functional genomics analysis to integrate genome-wide differential mRNA, miRNA, and protein expression as a function of human T lymphocyte activation and time. We surveyed expression of 420 human miRNAs in parallel with genome-wide mRNA expression. We identified a unique signature of 71 differentially expressed miRNAs, 57 of which were previously not known as regulators of immune activation. The majority of miRNAs are upregulated, mRNA expression of these target genes is downregulated, and this is a function of binding multiple miRNAs (combinatorial targeting). Our data reveal that consideration of this complex signature, rather than single miRNAs, is necessary to construct a full picture of miRNA-mediated regulation. Molecular network mapping of miRNA targets revealed the regulation of activation-induced immune signaling. In contrast, pathways populated by genes that are not miRNA targets are enriched for metabolism and biosynthesis. Finally, we specifically validated miR-155 (known) and miR-221 (novel in T lymphocytes) using locked nucleic acid inhibitors. Inhibition of these two highly upregulated miRNAs in CD4(+) T cells was shown to increase proliferation by removing suppression of four target genes linked to proliferation and survival. Thus, multiple lines of evidence link top functional networks directly to T lymphocyte immunity, underlining the value of mapping global gene, protein, and miRNA expression.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

