Flexibility of the C-terminal, or CII, ring of KaiC governs the rhythm of the circadian clock of cyanobacteria
- PMID: 21788479
- PMCID: PMC3167551
- DOI: 10.1073/pnas.1104221108
Flexibility of the C-terminal, or CII, ring of KaiC governs the rhythm of the circadian clock of cyanobacteria
Abstract
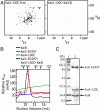
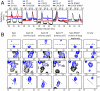
In the cyanobacterial circadian oscillator, KaiA and KaiB alternately stimulate autophosphorylation and autodephosphorylation of KaiC with a periodicity of approximately 24 h. KaiA activates autophosphorylation by selectively capturing the A loops of KaiC in their exposed positions. The A loops and sites of phosphorylation, residues S431 and T432, are located in the CII ring of KaiC. We find that the flexibility of the CII ring governs the rhythm of KaiC autophosphorylation and autodephosphorylation and is an example of dynamics-driven protein allostery. KaiA-induced autophosphorylation requires flexibility of the CII ring. In contrast, rigidity is required for KaiC-KaiB binding, which induces a conformational change in KaiB that enables it to sequester KaiA by binding to KaiA's linker. Autophosphorylation of the S431 residues around the CII ring stabilizes the CII ring, making it rigid. In contrast, autophosphorylation of the T432 residues offsets phospho-S431-induced rigidity to some extent. In the presence of KaiA and KaiB, the dynamic states of the CII ring of KaiC executes the following circadian rhythm: CII STflexible → CIISpTflexible → CIIpSpTrigid → CIIpSTvery-rigid → CIISTflexible. Apparently, these dynamic states govern the pattern of phosphorylation, ST → SpT → pSpT → pST → ST. CII-CI ring-on-ring stacking is observed when the CII ring is rigid, suggesting a mechanism through which the ATPase activity of the CI ring is rhythmically controlled. SasA, a circadian clock-output protein, binds to the CI ring. Thus, rhythmic ring stacking may also control clock-output pathways.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Comment in
-
Dynamic fluctuations lubricate the circadian clock.Proc Natl Acad Sci U S A. 2011 Aug 30;108(35):14377-8. doi: 10.1073/pnas.1111105108. Epub 2011 Aug 23. Proc Natl Acad Sci U S A. 2011. PMID: 21873208 Free PMC article. No abstract available.
References
-
- Albrecht U, editor. The Circadian Clock. New York: Springer; 2010.
-
- Dunlap JC, Loros JJ, DeCoursey PJ, editors. Chronobiology: Biological Timekeeping. Sunderland, MA: Sinauer; 2004.
-
- Ditty JL, Mackey SR, Johnson CH, editors. Bacterial Circadian Programs. Heidelberg: Springer; 2009.
-
- Ishiura M, et al. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

