Using an uncertainty-coding matrix in Bayesian regression models for haplotype-specific risk detection in family association studies
- PMID: 21789192
- PMCID: PMC3137600
- DOI: 10.1371/journal.pone.0021890
Using an uncertainty-coding matrix in Bayesian regression models for haplotype-specific risk detection in family association studies
Abstract
Haplotype association studies based on family genotype data can provide more biological information than single marker association studies. Difficulties arise, however, in the inference of haplotype phase determination and in haplotype transmission/non-transmission status. Incorporation of the uncertainty associated with haplotype inference into regression models requires special care. This task can get even more complicated when the genetic region contains a large number of haplotypes. To avoid the curse of dimensionality, we employ a clustering algorithm based on the evolutionary relationship among haplotypes and retain for regression analysis only the ancestral core haplotypes identified by it. To integrate the three sources of variation, phase ambiguity, transmission status and ancestral uncertainty, we propose an uncertainty-coding matrix which combines these three types of variability simultaneously. Next we evaluate haplotype risk with the use of such a matrix in a Bayesian conditional logistic regression model. Simulation studies and one application, a schizophrenia multiplex family study, are presented and the results are compared with those from other family based analysis tools such as FBAT. Our proposed method (Bayesian regression using uncertainty-coding matrix, BRUCM) is shown to perform better and the implementation in R is freely available.
Conflict of interest statement
Figures

 = 1.2 (1st column), 1.5 (2nd column) and 2.0 (3rd column). The first row contains posterior mean effects of
= 1.2 (1st column), 1.5 (2nd column) and 2.0 (3rd column). The first row contains posterior mean effects of  , the second is for its bias, and the last is for the posterior probability of susceptibility
, the second is for its bias, and the last is for the posterior probability of susceptibility  . Red plots correspond to the risk haplotypes.
. Red plots correspond to the risk haplotypes.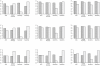
 = 1.2, 1.5, and 2.0, respectively. The three rows are simulations from additive (top), dominance (middle), and recessive models (bottom), respectively. The shaded bars in the left are under the hierarchical model with independent priors on regression coefficients, and the right bars contain results from FBAT.
= 1.2, 1.5, and 2.0, respectively. The three rows are simulations from additive (top), dominance (middle), and recessive models (bottom), respectively. The shaded bars in the left are under the hierarchical model with independent priors on regression coefficients, and the right bars contain results from FBAT.

 's (top two plots) and
's (top two plots) and  (bottom two plots) for schizophrenia study.
(bottom two plots) for schizophrenia study.References
-
- Zaykin D, Westfall P, Young S, Karnoub M, Wagner M, et al. Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Human Heredity. 2002;53:79–91. - PubMed
-
- Mensah FK, Gilthorpe MS, Davies CF, Keen LJ, Adamson PJ, et al. Haplotype uncertainty in association studies. Genetic Epidemiology. 2007;31:348–357. - PubMed
-
- Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, et al. Family-based tests for associating haplotypes with general phenotype data: Application to asthma genetics. Genetic Epidemiology. 2004;26:61–69. - PubMed
-
- Purcell S, Daly MJ, Sham PC. WHAP: haplotype-based association analysis. Bioinformatics. 2007;23:255–256. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

