Structure and assembly mechanism for heteromeric kainate receptors
- PMID: 21791290
- PMCID: PMC3145919
- DOI: 10.1016/j.neuron.2011.05.038
Structure and assembly mechanism for heteromeric kainate receptors
Abstract
Native glutamate receptor ion channels are tetrameric assemblies containing two or more different subunits. NMDA receptors are obligate heteromers formed by coassembly of two or three divergent gene families. While some AMPA and kainate receptors can form functional homomeric ion channels, the KA1 and KA2 subunits are obligate heteromers which function only in combination with GluR5-7. The mechanisms controlling glutamate receptor assembly involve an initial step in which the amino terminal domains (ATD) assemble as dimers. Here, we establish by sedimentation velocity that the ATDs of GluR6 and KA2 coassemble as a heterodimer of K(d) 11 nM, 32,000-fold lower than the K(d) for homodimer formation by KA2; we solve crystal structures for the GluR6/KA2 ATD heterodimer and heterotetramer assemblies. Using these structures as a guide, we perform a mutant cycle analysis to probe the energetics of assembly and show that high-affinity ATD interactions are required for biosynthesis of functional heteromeric receptors.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
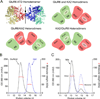
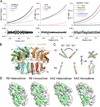
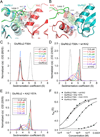

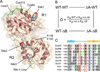
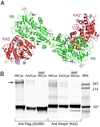
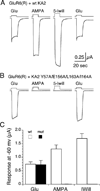
Comment in
-
How glutamate receptor subunits mix and match: details uncovered.Neuron. 2011 Jul 28;71(2):198-200. doi: 10.1016/j.neuron.2011.07.008. Neuron. 2011. PMID: 21791278
References
-
- Ayalon G, Segev E, Elgavish S, Stern-Bach Y. Two regions in the N-terminal domain of ionotropic glutamate receptor 3 form the subunit oligomerization interfaces that control subtype-specific receptor assembly. J Biol Chem. 2005;280:15053–15060. - PubMed
-
- Ayalon G, Stern-Bach Y. Functional assembly of AMPA and kainite receptors is mediated by several discrete protein-protein interactions. Neuron. 2001;31:103–113. - PubMed
-
- Brose N, Huntley GW, Stern-Bach Y, Sharma G, Morrison JH, Heinemann SF. Differential assembly of coexpressed glutamate receptor subunits in neurons of rat cerebral cortex. J Biol Chem. 1994;269:16780–16784. - PubMed
-
- Brown PH, Balbo A, Schuck P. Characterizing protein-protein interactions by sedimentation velocity analytical ultracentrifugation. Curr Protoc Immunol Chapter. 2008;18(Unit 18):15. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

