Reactivity of thioredoxin as a protein thiol-disulfide oxidoreductase
- PMID: 21793530
- PMCID: PMC3212873
- DOI: 10.1021/cr100006x
Reactivity of thioredoxin as a protein thiol-disulfide oxidoreductase
Figures


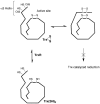
References
-
- Jensen KS, Hansen RE, Winther JR. Antioxid Redox Signal. 2009;11:1047. - PubMed
-
- Hatahet F, Ruddock LW. Antioxid Redox Signal. 2009;11:2807. - PubMed
-
- Jeong W, Yoon HW, Lee SR, Rhee SG. J Biol Chem. 2004;279:3142. - PubMed
-
- Powis G, Montfort WR. Annu Rev Pharmacol Toxicol. 2001;41:261. - PubMed
-
- Lillig CH, Holmgren A. Antioxid Redox Signal. 2007;9:25. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

