MicroRNA profiles in allograft tissues and paired urines associate with chronic allograft dysfunction with IF/TA
- PMID: 21794090
- PMCID: PMC3184368
- DOI: 10.1111/j.1600-6143.2011.03666.x
MicroRNA profiles in allograft tissues and paired urines associate with chronic allograft dysfunction with IF/TA
Abstract
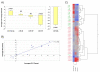
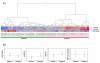
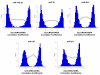
Despite the advances in immunosuppression, renal allograft attrition over time remains unabated due to chronic allograft dysfunction (CAD) with interstitial fibrosis (IF) and tubular atrophy (TA). We aimed to evaluate microRNA (miRNA) signatures in CAD with IF/TA and appraise correlation with paired urine samples and potential utility in prospective evaluation of graft function. MiRNA signatures were established between CAD with IF/TA versus normal allografts by microarray. Validation of the microarray results and prospective evaluation of urine samples was performed using real-time quantitative-PCR (RT-qPCR). Fifty-six miRNAs were identified in samples with CAD-IF/TA. Five miRNAs were selected for further validation based on array fold change, p-value and in silico predicted mRNA targets. We confirmed the differential expression of these five miRNAs by RT-qPCR using an independent set of samples. Differential expression was detected for miR-142-3p, miR-204, miR-107 and miR-211 (p < 0.001) and miR-32 (p < 0.05). Furthermore, differential expression of miR-142-3p (p < 0.01), miR-204 (p < 0.01) and miR-211 (p < 0.05) was also observed between patient groups in urine samples. A characteristic miRNA signature for IF/TA that correlates with paired urine samples was identified. These results support the potential use of miRNAs as noninvasive markers of IF/TA and for monitoring graft function.
©2011 The Authors Journal compilation©2011 The American Society of Transplantation and the American Society of Transplant Surgeons.
Figures



References
-
- Racusen LC, Solez K, Colvin RB, Bonsib SM, Castro MC, Cavallo T, et al. The Banff 97 working classification of renal allograft pathology. Kidney Int. 1999;55(2):713–723. - PubMed
-
- Solez K, Colvin RB, Racusen LC, Haas M, Sis B, Mengel M, et al. Banff 07 classification of renal allograft pathology: updates and future directions. Am.J.Transplant. 2008;8(4):753–760. - PubMed
-
- Schold JD, Kaplan B. The elephant in the room: failings of current clinical endpoints in kidney transplantation. Am.J.Transplant. 2010;10(5):1163–1166. - PubMed
-
- Poggio ED, Batty DS, Flechner SM. Evaluation of renal function in transplantation. Transplantation. 2007;84(2):131–136. - PubMed
-
- Manotham K, Booranalertpaisarn V, Eiam-Ong S, Chusil S, Praditpornsilpa K, Tungsanga K. Accurately simple estimation of glomerular filtration rate in kidney transplant patients. Transplant.Proc. 2002;34(4):1148–1151. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

