Transcriptional modulation induced by ionizing radiation: p53 remains a central player
- PMID: 21795128
- PMCID: PMC5528315
- DOI: 10.1016/j.molonc.2011.06.004
Transcriptional modulation induced by ionizing radiation: p53 remains a central player
Abstract
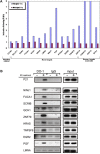
The cellular response to DNA damage is vital for maintaining genomic stability and preventing undue cell death or cancer formation. The DNA damage response (DDR), most robustly mobilized by double-strand breaks (DSBs), rapidly activates an extensive signaling network that affects numerous cellular systems, leading to cell survival or programmed cell death. A major component of the DDR is the widespread modulation of gene expression. We analyzed together six datasets that probed transcriptional responses to ionizing radiation (IR) - our novel experimental data and 5 published datasets - to elucidate the scope of this response and identify its gene targets. According to the mRNA expression profiles we recorded from 5 cancerous and non-cancerous human cell lines after exposure to 5 Gy of IR, most of the responses were cell line-specific. Computational analysis identified significant enrichment for p53 target genes and cell cycle-related pathways among groups of up-regulated and down-regulated genes, respectively. Computational promoter analysis of the six datasets disclosed that a statistically significant number of the induced genes contained p53 binding site signatures. p53-mediated regulation had previously been documented for subsets of these gene groups, making our lists a source of novel potential p53 targets. Real-time qPCR and chromatin immunoprecipitation (ChIP) assays validated the IR-induced p53-dependent induction and p53 binding to the respective promoters of 11 selected genes. Our results demonstrate the power of a combined computational and experimental approach to identify new transcriptional targets in the DNA damage response network.
Copyright © 2011 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures


References
-
- Adachi, N. , Nomoto, M. , 2000. Cell-cycle regulation of the DNA topoisomerase IIalpha promoter is mediated by proximal CCAAT boxes: possible involvement of acetylation. Gene. 245, (1) 49–57. - PubMed
-
- Akerman, G.S. , Rosenzweig, B.A. , 2005. "Alterations in gene expression profiles and the DNA-damage response in ionizing radiation-exposed TK6 cells.". Environ. Mol. Mutagen.. 45, (2–3) 188–205. - PubMed
-
- Alderson, M.R. , Smith, C.A. , 1994. Molecular and biological characterization of human 4-1BB and its ligand. Eur. J. Immunol.. 24, (9) 2219–2227. - PubMed
-
- Amundson, S.A. , Bittner, M. , 1999. Fluorescent cDNA microarray hybridization reveals complexity and heterogeneity of cellular genotoxic stress responses. Oncogene. 18, (24) 3666–3672. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

