Epstein-Barr virus nuclear antigen 1 replication and segregation functions in nasopharyngeal carcinoma cell lines
- PMID: 21795327
- PMCID: PMC3196394
- DOI: 10.1128/JVI.05293-11
Epstein-Barr virus nuclear antigen 1 replication and segregation functions in nasopharyngeal carcinoma cell lines
Abstract
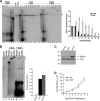
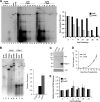
Nasopharyngeal carcinomas (NPC) are usually Epstein-Barr virus (EBV) positive, but, with the exception of C666-1 cells, these cells lose the EBV genomes when grown in culture. Maintenance of EBV requires the viral EBV nuclear antigen 1 (EBNA1) protein, which ensures the replication and mitotic segregation of the genomes through interactions with OriP. Here we compare the abilities of C666-1 and NPC cells that have lost EBV genomes to replicate and segregate OriP plasmids. We found that either cell line can replicate and maintain OriP plasmids for extended periods under conditions where low levels of EBNA1 are expressed but that high EBNA1 levels selectively interfered with mitotic segregation.
Figures


Similar articles
-
Structural Basis for Cooperative Binding of EBNA1 to the Epstein-Barr Virus Dyad Symmetry Minimal Origin of Replication.J Virol. 2019 Sep 30;93(20):e00487-19. doi: 10.1128/JVI.00487-19. Print 2019 Oct 15. J Virol. 2019. PMID: 31142669 Free PMC article.
-
Structural and Functional Basis for an EBNA1 Hexameric Ring in Epstein-Barr Virus Episome Maintenance.J Virol. 2017 Sep 12;91(19):e01046-17. doi: 10.1128/JVI.01046-17. Print 2017 Oct 1. J Virol. 2017. PMID: 28701406 Free PMC article.
-
Carcinoma-risk variant of EBNA1 deregulates Epstein-Barr Virus episomal latency.Oncotarget. 2017 Jan 31;8(5):7248-7264. doi: 10.18632/oncotarget.14540. Oncotarget. 2017. PMID: 28077791 Free PMC article.
-
Replication licensing of the EBV oriP minichromosome.Curr Top Microbiol Immunol. 2001;258:13-33. doi: 10.1007/978-3-642-56515-1_2. Curr Top Microbiol Immunol. 2001. PMID: 11443858 Review.
-
EBNA1.Curr Top Microbiol Immunol. 2015;391:3-34. doi: 10.1007/978-3-319-22834-1_1. Curr Top Microbiol Immunol. 2015. PMID: 26428370 Review.
Cited by
-
Molecular pathological study of the human nasopharyngeal carcinoma CNE3 cell line.Oncol Lett. 2013 Oct;6(4):980-984. doi: 10.3892/ol.2013.1513. Epub 2013 Aug 5. Oncol Lett. 2013. PMID: 24137449 Free PMC article.
-
Understanding the interplay between host immunity and Epstein-Barr virus in NPC patients.Emerg Microbes Infect. 2015 Mar;4(3):e20. doi: 10.1038/emi.2015.20. Epub 2015 Mar 25. Emerg Microbes Infect. 2015. PMID: 26038769 Free PMC article. Review.
-
Transcriptome-wide analysis of alternative RNA splicing events in Epstein-Barr virus-associated gastric carcinomas.PLoS One. 2017 May 11;12(5):e0176880. doi: 10.1371/journal.pone.0176880. eCollection 2017. PLoS One. 2017. PMID: 28493890 Free PMC article.
-
Epstein-Barr Virus Nuclear Antigen 1 Recruits Cyclophilin A to Facilitate the Replication of Viral DNA Genome.Front Microbiol. 2019 Dec 13;10:2879. doi: 10.3389/fmicb.2019.02879. eCollection 2019. Front Microbiol. 2019. PMID: 31921057 Free PMC article.
-
Latent Membrane Protein 1 Is a Novel Determinant of Epstein-Barr Virus Genome Persistence and Reactivation.mSphere. 2017 Nov 8;2(6):e00453-17. doi: 10.1128/mSphereDirect.00453-17. eCollection 2017 Nov-Dec. mSphere. 2017. PMID: 29134204 Free PMC article.
References
-
- Cheung S. T., et al. 1999. Nasopharyngeal carcinoma cell line (C666-1) consistently harbouring Epstein-Barr virus. Int. J. Cancer 83:121–126 - PubMed
-
- de Jesus O., et al. 2003. Updated Epstein-Barr virus (EBV) DNA sequence and analysis of a promoter for the BART (CST, BARF0) RNAs of EBV. J. Gen. Virol. 84:1443–1450 - PubMed
-
- Frappier L. 2010. EBNA1 in viral DNA replication and persistence, p. 37–59 In Robertson E. S. (ed.), Epstein-Barr virus: latency and transformation. Caister Academic Press, Norwich, United Kingdom
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

