Reassortment events among swine influenza A viruses in China: implications for the origin of the 2009 influenza pandemic
- PMID: 21795347
- PMCID: PMC3196454
- DOI: 10.1128/JVI.05262-11
Reassortment events among swine influenza A viruses in China: implications for the origin of the 2009 influenza pandemic
Abstract
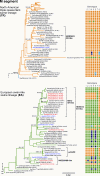
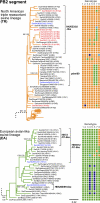
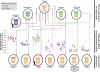
That pigs may play a pivotal role in the emergence of pandemic influenza was indicated by the recent H1N1/2009 human pandemic, likely caused by a reassortant between viruses of the American triple-reassortant (TR) and Eurasian avian-like (EA) swine influenza lineages. As China has the largest human and pig populations in the world and is the only place where both TR and EA viruses have been reported to cocirculate, it is potentially the source of the H1N1/2009 pandemic virus. To examine this, the genome sequences of 405 swine influenza viruses from China were analyzed. Thirty-six TR and EA reassortant viruses were identified before and after the occurrence of the pandemic. Several of these TR-EA reassortant viruses had genotypes with most segments having the same lineage origin as the segments of the H1N1/2009 pandemic virus. However, these viruses were generated from independent reassortment events throughout our survey period and were not associated with the current pandemic. One TR-EA reassortant, which is least similar to the pandemic virus, has persisted since 2007, while all the other variants appear to be transient. Despite frequent reassortment events between TR and EA lineage viruses in China, evidence for the genesis of the 2009 pandemic virus in pigs in this region is still absent.
Figures
References
-
- Brown I. H. 2000. The epidemiology and evolution of influenza viruses in pigs. Vet. Microbiol. 74:29–46 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

