Quantitative single cell monitoring of protein synthesis at subcellular resolution using fluorescently labeled tRNA
- PMID: 21795382
- PMCID: PMC3201886
- DOI: 10.1093/nar/gkr601
Quantitative single cell monitoring of protein synthesis at subcellular resolution using fluorescently labeled tRNA
Abstract
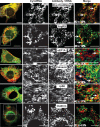
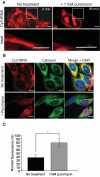
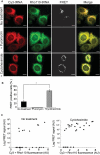
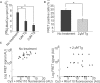
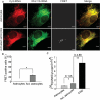
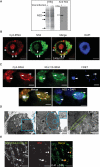
We have developed a novel technique of using fluorescent tRNA for translation monitoring (FtTM). FtTM enables the identification and monitoring of active protein synthesis sites within live cells at submicron resolution through quantitative microscopy of transfected bulk uncharged tRNA, fluorescently labeled in the D-loop (fl-tRNA). The localization of fl-tRNA to active translation sites was confirmed through its co-localization with cellular factors and its dynamic alterations upon inhibition of protein synthesis. Moreover, fluorescence resonance energy transfer (FRET) signals, generated when fl-tRNAs, separately labeled as a FRET pair occupy adjacent sites on the ribosome, quantitatively reflect levels of protein synthesis in defined cellular regions. In addition, FRET signals enable detection of intra-populational variability in protein synthesis activity. We demonstrate that FtTM allows quantitative comparison of protein synthesis between different cell types, monitoring effects of antibiotics and stress agents, and characterization of changes in spatial compartmentalization of protein synthesis upon viral infection.
Figures