Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1
- PMID: 21795385
- PMCID: PMC3202282
- DOI: 10.1101/gr.121145.111
Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1
Abstract
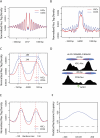
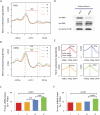
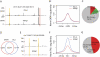
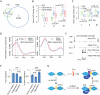
Enhancers of transcription activate transcription via binding of sequence-specific transcription factors to their target sites in chromatin. In this report, we identify GATA1-bound distal sites genome-wide and find a global reorganization of the nucleosomes at these potential enhancers during differentiation of hematopoietic stem cells (HSCs) to erythrocytes. We show that the catalytic subunit BRG1 of BAF complexes localizes to these distal sites during differentiation and generates a longer nucleosome linker region surrounding the GATA1 sites by shifting the flanking nucleosomes away. Intriguingly, we find that the nucleosome shifting specifically facilitates binding of TAL1 but not GATA1 and is linked to subsequent transcriptional regulation of target genes.
Figures

References
-
- Armstrong JA, Bieker JJ, Emerson BM 1998. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 95: 93–104 - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 - PubMed
-
- Boyes J, Omichinski J, Clark D, Pikaart M, Felsenfeld G 1998. Perturbation of nucleosome structure by the erythroid transcription factor GATA-1. J Mol Biol 279: 529–544 - PubMed
-
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, et al. 2000. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell 6: 1287–1295 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
