Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration
- PMID: 21795386
- PMCID: PMC3202283
- DOI: 10.1101/gr.125088.111
Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration
Abstract
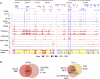
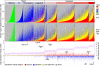
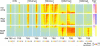
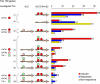
Interplays among lineage-specific nuclear proteins, chromatin modifying enzymes, and the basal transcription machinery govern cellular differentiation, but their dynamics of action and coordination with transcriptional control are not fully understood. Alterations in chromatin structure appear to establish a permissive state for gene activation at some loci, but they play an integral role in activation at other loci. To determine the predominant roles of chromatin states and factor occupancy in directing gene regulation during differentiation, we mapped chromatin accessibility, histone modifications, and nuclear factor occupancy genome-wide during mouse erythroid differentiation dependent on the master regulatory transcription factor GATA1. Notably, despite extensive changes in gene expression, the chromatin state profiles (proportions of a gene in a chromatin state dominated by activating or repressive histone modifications) and accessibility remain largely unchanged during GATA1-induced erythroid differentiation. In contrast, gene induction and repression are strongly associated with changes in patterns of transcription factor occupancy. Our results indicate that during erythroid differentiation, the broad features of chromatin states are established at the stage of lineage commitment, largely independently of GATA1. These determine permissiveness for expression, with subsequent induction or repression mediated by distinctive combinations of transcription factors.
Figures


References
-
- Barton MC, Crowe AJ 2001. Chromatin alteration, transcription and replication: What's the opening line to the story? Oncogene 20: 3094–3099 - PubMed
-
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- R01 HG004037/HG/NHGRI NIH HHS/United States
- R01HG002238/HG/NHGRI NIH HHS/United States
- R01HG004718/HG/NHGRI NIH HHS/United States
- R01DK065806/DK/NIDDK NIH HHS/United States
- R01DK58044/DK/NIDDK NIH HHS/United States
- RC2HG005639/HG/NHGRI NIH HHS/United States
- R01 HG004718/HG/NHGRI NIH HHS/United States
- RC2 HG005573/HG/NHGRI NIH HHS/United States
- R01 DK058044/DK/NIDDK NIH HHS/United States
- R01 DK065806/DK/NIDDK NIH HHS/United States
- RC2HG005573/HG/NHGRI NIH HHS/United States
- R01 DK054937/DK/NIDDK NIH HHS/United States
- RC1 HG005334/HG/NHGRI NIH HHS/United States
- R01 HG002238/HG/NHGRI NIH HHS/United States
- RC2 HG005639/HG/NHGRI NIH HHS/United States
- RC1HG005334/HG/NHGRI NIH HHS/United States
- R01DK54937/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
