A proteogenomic analysis of Anopheles gambiae using high-resolution Fourier transform mass spectrometry
- PMID: 21795387
- PMCID: PMC3205572
- DOI: 10.1101/gr.127951.111
A proteogenomic analysis of Anopheles gambiae using high-resolution Fourier transform mass spectrometry
Abstract
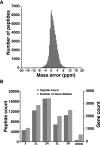
Anopheles gambiae is a major mosquito vector responsible for malaria transmission, whose genome sequence was reported in 2002. Genome annotation is a continuing effort, and many of the approximately 13,000 genes listed in VectorBase for Anopheles gambiae are predictions that have still not been validated by any other method. To identify protein-coding genes of An. gambiae based on its genomic sequence, we carried out a deep proteomic analysis using high-resolution Fourier transform mass spectrometry for both precursor and fragment ions. Based on peptide evidence, we were able to support or correct more than 6000 gene annotations including 80 novel gene structures and about 500 translational start sites. An additional validation by RT-PCR and cDNA sequencing was successfully performed for 105 selected genes. Our proteogenomic analysis led to the identification of 2682 genome search-specific peptides. Numerous cases of encoded proteins were documented in regions annotated as intergenic, introns, or untranslated regions. Using a database created to contain potential splice sites, we also identified 35 novel splice junctions. This is a first report to annotate the An. gambiae genome using high-accuracy mass spectrometry data as a complementary technology for genome annotation.
Figures



References
-
- Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S 2008. Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320: 938–941 - PubMed
-
- Brunner E, Ahrens CH, Mohanty S, Baetschmann H, Loevenich S, Potthast F, Deutsch EW, Panse C, de Lichtenberg U, Rinner O, et al. 2007. A high-quality catalog of the Drosophila melanogaster proteome. Nat Biotechnol 25: 576–583 - PubMed
-
- Choudhary JS, Blackstock WP, Creasy DM, Cottrell JS 2001. Matching peptide mass spectra to EST and genomic DNA databases. Trends Biotechnol 19: S17–S22 - PubMed
-
- Driessen HP, de Jong WW, Tesser GI, Bloemendal H 1985. The mechanism of N-terminal acetylation of proteins. CRC Crit Rev Biochem 18: 281–325 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
