GSK3β inactivation induces apoptosis of leukemia cells by repressing the function of c-Myb
- PMID: 21795403
- PMCID: PMC3172276
- DOI: 10.1091/mbc.E11-06-0483
GSK3β inactivation induces apoptosis of leukemia cells by repressing the function of c-Myb
Abstract
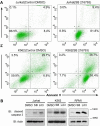
Glycogen synthase kinase 3β (GSK3β) regulates diverse physiological processes, including metabolism, development, oncogenesis, and neuroprotection. GSK3β kinase activity has been reported to be critical for various types of cancer cells, but the mechanism has remained elusive. In this study we examine the mechanism by which GSK3β regulates the survival of leukemia cells. We demonstrate that upon GSK3β kinase inhibition different types of leukemia cells show severe proliferation defects as a result of apoptosis. The transcription factor c-Myb is found to be the main target of GSK3β inhibition in cell survival. GSK3β inactivation reduces the expression of c-Myb by promoting its ubiquitination-mediated degradation, thereby inhibiting the expression of c-Myb-dependent antiapoptotic genes Bcl2 and survivin. Coimmunoprecipitation, reporter assays, chromatin immunoprecipitation, and knockdown studies show that c-Myb needs to interact and cooperate with transcription factor LEF-1 in the activation of Bcl2 and survivin and that both transcription factors are required for cell survival. These data reveal an as-yet-unknown mechanism by which GSK3β controls cell survival.
Figures


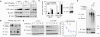


References
-
- Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. - PubMed
-
- Beug H, von Kirchbach A, Doderlein G, Conscience JF, Graf T. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell. 1979;18:375–390. - PubMed
-
- Corradini F, Cesi V, Bartella V, Pani E, Bussolari R, Candini O, Calabretta B. Enhanced proliferative potential of hematopoietic cells expressing degradation-resistant c-Myb mutants. J Biol Chem. 2005;280:30254–30262. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

