Genomic species are ecological species as revealed by comparative genomics in Agrobacterium tumefaciens
- PMID: 21795751
- PMCID: PMC3163468
- DOI: 10.1093/gbe/evr070
Genomic species are ecological species as revealed by comparative genomics in Agrobacterium tumefaciens
Abstract
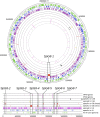
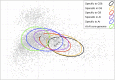
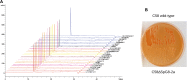
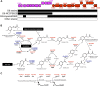
The definition of bacterial species is based on genomic similarities, giving rise to the operational concept of genomic species, but the reasons of the occurrence of differentiated genomic species remain largely unknown. We used the Agrobacterium tumefaciens species complex and particularly the genomic species presently called genomovar G8, which includes the sequenced strain C58, to test the hypothesis of genomic species having specific ecological adaptations possibly involved in the speciation process. We analyzed the gene repertoire specific to G8 to identify potential adaptive genes. By hybridizing 25 strains of A. tumefaciens on DNA microarrays spanning the C58 genome, we highlighted the presence and absence of genes homologous to C58 in the taxon. We found 196 genes specific to genomovar G8 that were mostly clustered into seven genomic islands on the C58 genome-one on the circular chromosome and six on the linear chromosome-suggesting higher plasticity and a major adaptive role of the latter. Clusters encoded putative functional units, four of which had been verified experimentally. The combination of G8-specific functions defines a hypothetical species primary niche for G8 related to commensal interaction with a host plant. This supports that the G8 ancestor was able to exploit a new ecological niche, maybe initiating ecological isolation and thus speciation. Searching genomic data for synapomorphic traits is a powerful way to describe bacterial species. This procedure allowed us to find such phenotypic traits specific to genomovar G8 and thus propose a Latin binomial, Agrobacterium fabrum, for this bona fide genomic species.
Figures


References
-
- Achtman M, Wagner M. Microbial diversity and the genetic nature of microbial species. Nat Rev Microbiol. 2008;6:439. - PubMed
-
- Bouzar H, Jones J. Agrobacterium larrymoorei sp. nov., a pathogen isolated from aerial tumours of Ficus benjamina. Int J Syst Evol Microbiol. 2001;51:1023–1026. - PubMed
-
- Braun V, Mahren S, Sauter A. Gene regulation by transmembrane signaling. Biometals. 2006;19:103–113. - PubMed
-
- Breese K, Fuchs G. 4-Hydroxybenzoyl-CoA reductase (dehydroxylating) from the denitrifying bacterium Thauera aromatica—prosthetic groups, electron donor, and genes of a member of the molybdenum-flavin-iron-sulfur proteins. Eur J Biochem. 1998;251:916–923. - PubMed

