Transforming binding affinities from three dimensions to two with application to cadherin clustering
- PMID: 21796210
- PMCID: PMC3167384
- DOI: 10.1038/nature10183
Transforming binding affinities from three dimensions to two with application to cadherin clustering
Abstract
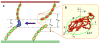
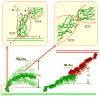
Membrane-bound receptors often form large assemblies resulting from binding to soluble ligands, cell-surface molecules on other cells and extracellular matrix proteins. For example, the association of membrane proteins with proteins on different cells (trans-interactions) can drive the oligomerization of proteins on the same cell (cis-interactions). A central problem in understanding the molecular basis of such phenomena is that equilibrium constants are generally measured in three-dimensional solution and are thus difficult to relate to the two-dimensional environment of a membrane surface. Here we present a theoretical treatment that converts three-dimensional affinities to two dimensions, accounting directly for the structure and dynamics of the membrane-bound molecules. Using a multiscale simulation approach, we apply the theory to explain the formation of ordered, junction-like clusters by classical cadherin adhesion proteins. The approach features atomic-scale molecular dynamics simulations to determine interdomain flexibility, Monte Carlo simulations of multidomain motion and lattice simulations of junction formation. A finding of general relevance is that changes in interdomain motion on trans-binding have a crucial role in driving the lateral, cis-, clustering of adhesion receptors.
Figures


References
-
- Aplin AE, Howe AK, Juliano RL. Cell adhesion molecules, signal transduction and cell growth. Curr Opin Cell Biol. 1999;11:737–744. - PubMed
-
- Aricescu AR, Jones EY. Immunoglobulin superfamily cell adhesion molecules: zippers and signals. Curr Opin Cell Biol. 2007;19:543–550. - PubMed
-
- Dustin ML, Bromley SK, Davis MM, Zhu C. Identification of self through two-dimensional chemistry and synapses. Annu Rev Cell Dev Biol. 2001;17:133–157. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

