Pathway specific gene expression profiling reveals oxidative stress genes potentially regulated by transcription co-activator LEDGF/p75 in prostate cancer cells
- PMID: 21796653
- PMCID: PMC3227744
- DOI: 10.1002/pros.21463
Pathway specific gene expression profiling reveals oxidative stress genes potentially regulated by transcription co-activator LEDGF/p75 in prostate cancer cells
Abstract
Background: Lens epithelium-derived growth factor p75 (LEDGF/p75) is a stress survival transcription co-activator and autoantigen that is overexpressed in tumors, including prostate cancer (PCa). This oncoprotein promotes resistance to cell death induced by oxidative stress and chemotherapy by mechanisms that remain unclear. To get insights into these mechanisms we identified candidate target stress genes of LEDGF/p75 using pathway-specific gene expression profiling in PCa cells.
Methods: A "Human oxidative stress and antioxidant defense" qPCR array was used to identify genes exhibiting significant expression changes in response to knockdown or overexpression of LEDGF/p75 in PC-3 cells. Validation of array results was performed by additional qPCR and immunoblotting.
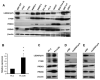
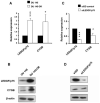
Results: Cytoglobin (CYGB), Phosphoinositide-binding protein PIP3-E/IPCEF-1, superoxidase dismutase 3 (SOD3), thyroid peroxidase (TPO), and albumin (ALB) exhibited significant transcript down- and up-regulation in response to LEDGF/p75 knockdown and overexpression, respectively. CYGB gene was selected for further validation based on its emerging role as a stress oncoprotein in human malignancies. In light of previous reports indicating that LEDGF/p75 regulates peroxiredoxin 6 (PRDX6), and that PRDXs exhibit differential expression in PCa, we also examined the relationship between these proteins in PCa cells. Our validation data revealed that changes in LEDGF/p75 transcript and protein expression in PCa cells closely paralleled those of CYGB, but not those of the PRDXs.
Conclusions: Our study identifies CYGB and other genes as stress genes potentially regulated by LEDGF/p75 in PCa cells, and provides a rationale for investigating their role in PCa and in promoting resistance to chemotherapy- and oxidative stress-induced cell death.
Copyright © 2011 Wiley Periodicals, Inc.
Figures
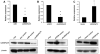



Similar articles
-
LEDGF/p75 Overexpression Attenuates Oxidative Stress-Induced Necrosis and Upregulates the Oxidoreductase ERP57/PDIA3/GRP58 in Prostate Cancer.PLoS One. 2016 Jan 15;11(1):e0146549. doi: 10.1371/journal.pone.0146549. eCollection 2016. PLoS One. 2016. PMID: 26771192 Free PMC article.
-
Targeting the stress oncoprotein LEDGF/p75 to sensitize chemoresistant prostate cancer cells to taxanes.Oncotarget. 2017 Apr 11;8(15):24915-24931. doi: 10.18632/oncotarget.15323. Oncotarget. 2017. PMID: 28212536 Free PMC article.
-
The LEDGF/p75 Integrase Binding Domain Interactome Contributes to the Survival, Clonogenicity, and Tumorsphere Formation of Docetaxel-Resistant Prostate Cancer Cells.Cells. 2021 Oct 12;10(10):2723. doi: 10.3390/cells10102723. Cells. 2021. PMID: 34685704 Free PMC article.
-
Role of LEDGF/p75 (PSIP1) in oncogenesis. Insights in molecular mechanism and therapeutic potential.Biochim Biophys Acta Rev Cancer. 2025 Feb;1880(1):189248. doi: 10.1016/j.bbcan.2024.189248. Epub 2024 Dec 18. Biochim Biophys Acta Rev Cancer. 2025. PMID: 39701326 Review.
-
LEDGF/p75-mediated chemoresistance of mixed-lineage leukemia involves cell survival pathways and super enhancer activators.Cancer Gene Ther. 2022 Feb;29(2):133-140. doi: 10.1038/s41417-021-00319-3. Epub 2021 Apr 1. Cancer Gene Ther. 2022. PMID: 33795806 Review.
Cited by
-
Unlike Its Paralog LEDGF/p75, HRP-2 Is Dispensable for MLL-R Leukemogenesis but Important for Leukemic Cell Survival.Cells. 2021 Jan 19;10(1):192. doi: 10.3390/cells10010192. Cells. 2021. PMID: 33477970 Free PMC article.
-
Twenty years of research on the DFS70/LEDGF autoantibody-autoantigen system: many lessons learned but still many questions.Auto Immun Highlights. 2020 Feb 3;11(1):3. doi: 10.1186/s13317-020-0126-4. Auto Immun Highlights. 2020. PMID: 32127038 Free PMC article. Review.
-
Oncogenic human papillomaviruses activate the tumor-associated lens epithelial-derived growth factor (LEDGF) gene.PLoS Pathog. 2014 Mar 6;10(3):e1003957. doi: 10.1371/journal.ppat.1003957. eCollection 2014 Mar. PLoS Pathog. 2014. PMID: 24604027 Free PMC article.
-
Cytoglobin in tumor hypoxia: novel insights into cancer suppression.Tumour Biol. 2014 Jul;35(7):6207-19. doi: 10.1007/s13277-014-1992-z. Epub 2014 May 10. Tumour Biol. 2014. PMID: 24816917 Review.
-
Impact of EcSOD Perturbations in Cancer Progression.Antioxidants (Basel). 2021 Jul 29;10(8):1219. doi: 10.3390/antiox10081219. Antioxidants (Basel). 2021. PMID: 34439467 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

