Identifying protein variants with cross-reactive aptamer arrays
- PMID: 21796750
- PMCID: PMC3454492
- DOI: 10.1002/cbic.201100046
Identifying protein variants with cross-reactive aptamer arrays
Figures
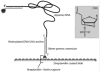
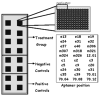


Similar articles
-
Isolation of Peptide aptamers to target protein function.Methods Mol Biol. 2009;535:333-60. doi: 10.1007/978-1-59745-557-2_19. Methods Mol Biol. 2009. PMID: 19377987
-
Prediction of aptamer-protein interacting pairs using an ensemble classifier in combination with various protein sequence attributes.BMC Bioinformatics. 2016 May 31;17(1):225. doi: 10.1186/s12859-016-1087-5. BMC Bioinformatics. 2016. PMID: 27245069 Free PMC article.
-
Development of an Aptamer-Based Lateral Flow Assay for the Detection of C-Reactive Protein Using Microarray Technology as a Prescreening Platform.ACS Comb Sci. 2020 Nov 9;22(11):617-629. doi: 10.1021/acscombsci.0c00080. Epub 2020 Sep 7. ACS Comb Sci. 2020. PMID: 32894679
-
Peptide aptamers as guides for small-molecule drug discovery.Drug Discov Today. 2006 Apr;11(7-8):334-41. doi: 10.1016/j.drudis.2006.02.007. Drug Discov Today. 2006. PMID: 16580975 Review.
-
Peptide aptamers: development and applications.Curr Top Med Chem. 2015;15(12):1082-101. doi: 10.2174/1568026615666150413153143. Curr Top Med Chem. 2015. PMID: 25866267 Free PMC article. Review.
Cited by
-
A differential fluorescent receptor for nucleic acid analysis.Chembiochem. 2014 Jan 24;15(2):228-31. doi: 10.1002/cbic.201300657. Epub 2013 Dec 11. Chembiochem. 2014. PMID: 24339354 Free PMC article.
-
Array-based "Chemical Nose" Sensing in Diagnostics and Drug Discovery.Angew Chem Int Ed Engl. 2019 Apr 8;58(16):5190-5200. doi: 10.1002/anie.201809607. Epub 2019 Feb 20. Angew Chem Int Ed Engl. 2019. PMID: 30347522 Free PMC article. Review.
-
Next-generation sequencing as input for chemometrics in differential sensing routines.Angew Chem Int Ed Engl. 2015 May 18;54(21):6339-42. doi: 10.1002/anie.201501822. Epub 2015 Mar 31. Angew Chem Int Ed Engl. 2015. PMID: 25826754 Free PMC article.
-
Fingerprinting Non-Terran Biosignatures.Astrobiology. 2018 Jul;18(7):915-922. doi: 10.1089/ast.2017.1712. Epub 2018 Mar 8. Astrobiology. 2018. PMID: 29634318 Free PMC article.
-
Small molecule detection with aptamer based lateral flow assays: Applying aptamer-C-reactive protein cross-recognition for ampicillin detection.Sci Rep. 2018 Apr 4;8(1):5628. doi: 10.1038/s41598-018-23963-6. Sci Rep. 2018. PMID: 29618771 Free PMC article.
References
-
- Conroy PJ, Hearty S, Leonard P, O’Kennedy RJ. Semin Cell Dev Biol. 2009;20:10–26. - PubMed
- Luppa PB, Sokoll LJ, Chan DW. Clin Chim Acta. 2001;314:1–26. - PubMed
- Goodey A, Lavigne JJ, Savoy SM, Rodriguez MD, Curey T, Tsao A, Simmons G, Wright J, Yoo SJ, Sohn Y, Anslyn EV, Shear JB, Neikirk DP, McDevitt JT. J AM CHEM SOC. 2001;123:2559–2570. - PubMed
- Haab BB, Dunham MJ, Brown PO. Genome Biol. 2001;2 RESEARCH0004. - PMC - PubMed
-
- Miller JC, Zhou H, Kwekel J, Cavallo R, Burke J, Butler EB, Teh BS, Haab BB. Proteomics. 2003;3:56–63. - PubMed
-
- Lavigne J, Anslyn E. ANGEW CHEM INT EDIT. 2001;40:3119–3130. - PubMed
- Dickinson TA, White J, Kauer JS, Walt DR. Nature. 1996;382:697–700. - PubMed
- Albert KJ, Lewis NS, Schauer CL, Sotzing GA, Stitzel SE, Vaid TP, Walt DR. CHEM REV. 2000;100:2595–2626. - PubMed
- Lewis NS. Acc Chem Res. 2004;37:663–672. - PubMed
-
- Shangguan D, Cao Z, Meng L, Mallikaratchy P, Sefah K, Wang H, Li Y, Tan W. J PROTEOME RES. 2008;7:2133–2139. - PMC - PubMed
- Suslick K, Rakow N, Sen A. TETRAHEDRON. 2004;60:11133–11138.
- Stojanovic M, Green E, Semova S, Nikic D, Landry D. J AM CHEM SOC. 2003;125:6085–6089. - PubMed
- Fitter S, James R. J BIOL CHEM. 2005;280:34193–34201. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

