Molecular evolution of Azagny virus, a newfound hantavirus harbored by the West African pygmy shrew (Crocidura obscurior) in Côte d'Ivoire
- PMID: 21798050
- PMCID: PMC3163557
- DOI: 10.1186/1743-422X-8-373
Molecular evolution of Azagny virus, a newfound hantavirus harbored by the West African pygmy shrew (Crocidura obscurior) in Côte d'Ivoire
Abstract
Background: Tanganya virus (TGNV), the only shrew-associated hantavirus reported to date from sub-Saharan Africa, is harbored by the Therese's shrew (Crocidura theresae), and is phylogenetically distinct from Thottapalayam virus (TPMV) in the Asian house shrew (Suncus murinus) and Imjin virus (MJNV) in the Ussuri white-toothed shrew (Crocidura lasiura). The existence of myriad soricid-borne hantaviruses in Eurasia and North America would predict the presence of additional hantaviruses in sub-Saharan Africa, where multiple shrew lineages have evolved and diversified.
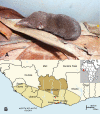
Methods: Lung tissues, collected in RNAlater®, from 39 Buettikofer's shrews (Crocidura buettikoferi), 5 Jouvenet's shrews (Crocidura jouvenetae), 9 West African pygmy shrews (Crocidura obscurior) and 21 African giant shrews (Crocidura olivieri) captured in Côte d'Ivoire during 2009, were systematically examined for hantavirus RNA by RT-PCR.
Results: A genetically distinct hantavirus, designated Azagny virus (AZGV), was detected in the West African pygmy shrew. Phylogenetic analysis of the S, M and L segments, using maximum-likelihood and Bayesian methods, under the GTR+I+Γ model of evolution, showed that AZGV shared a common ancestry with TGNV and was more closely related to hantaviruses harbored by soricine shrews than to TPMV and MJNV. That is, AZGV in the West African pygmy shrew, like TGNV in the Therese's shrew, did not form a monophyletic group with TPMV and MJNV, which were deeply divergent and basal to other rodent- and soricomorph-borne hantaviruses. Ancestral distributions of each hantavirus lineage, reconstructed using Mesquite 2.74, suggested that the common ancestor of all hantaviruses was most likely of Eurasian, not African, origin.
Conclusions: Genome-wide analysis of many more hantaviruses from sub-Saharan Africa are required to better understand how the biogeographic origin and radiation of African shrews might have contributed to, or have resulted from, the evolution of hantaviruses.
Figures

References
-
- Frame JD, Baldwin JM Jr, Gocke DJ, Troup JM. Lassa fever, a new virus disease of man from West Africa. I. Clinical description and pathological findings. Am J Trop Med Hyg. 1970;19:670–676. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

