Allelic heterogeneity and more detailed analyses of known loci explain additional phenotypic variation and reveal complex patterns of association
- PMID: 21798870
- PMCID: PMC3177649
- DOI: 10.1093/hmg/ddr328
Allelic heterogeneity and more detailed analyses of known loci explain additional phenotypic variation and reveal complex patterns of association
Abstract
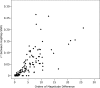
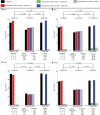
The identification of multiple signals at individual loci could explain additional phenotypic variance ('missing heritability') of common traits, and help identify causal genes. We examined gene expression levels as a model trait because of the large number of strong genetic effects acting in cis. Using expression profiles from 613 individuals, we performed genome-wide single nucleotide polymorphism (SNP) analyses to identify cis-expression quantitative trait loci (eQTLs), and conditional analysis to identify second signals. We examined patterns of association when accounting for multiple SNPs at a locus and when including additional SNPs from the 1000 Genomes Project. We identified 1298 cis-eQTLs at an approximate false discovery rate 0.01, of which 118 (9%) showed evidence of a second independent signal. For this subset of 118 traits, accounting for two signals resulted in an average 31% increase in phenotypic variance explained (Wilcoxon P< 0.0001). The association of SNPs with cis gene expression could increase, stay similar or decrease in significance when accounting for linkage disequilibrium with second signals at the same locus. Pairs of SNPs increasing in significance tended to have gene expression increasing alleles on opposite haplotypes, whereas pairs of SNPs decreasing in significance tended to have gene expression increasing alleles on the same haplotypes. Adding data from the 1000 Genomes Project showed that apparently independent signals could be potentially explained by a single association signal. Our results show that accounting for multiple variants at a locus will increase the variance explained in a substantial fraction of loci, but that allelic heterogeneity will be difficult to define without resequencing loci and functional work.
Figures




References
-
- Spencer C.C., Plagnol V., Strange A., Gardner M., Paisan-Ruiz C., Band G., Barker R.A., Bellenguez C., Bhatia K., Blackburn H., et al. Dissection of the genetics of Parkinson's disease identifies an additional association 5′ of SNCA and multiple associated haplotypes at 17q21. Hum. Mol. Genet. 2011;20:345–353. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

