Allergic asthma: influence of genetic and environmental factors
- PMID: 21799018
- PMCID: PMC3190897
- DOI: 10.1074/jbc.R110.197046
Allergic asthma: influence of genetic and environmental factors
Abstract
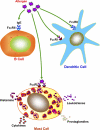
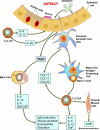
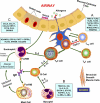
Allergic asthma is a chronic airway inflammatory disease in which exposure to allergens causes intermittent attacks of breathlessness, airway hyper-reactivity, wheezing, and coughing. Allergic asthma has been called a "syndrome" resulting from a complex interplay between genetic and environmental factors. Worldwide, >300 million individuals are affected by this disease, and in the United States alone, it is estimated that >35 million people, mostly children, suffer from asthma. Although animal models, linkage analyses, and genome-wide association studies have identified numerous candidate genes, a solid definition of allergic asthma has not yet emerged; however, such studies have contributed to our understanding of the multiple pathways to this syndrome. In contrast with animal models, in which T-helper 2 (T(H)2) cell response is the dominant feature, in human asthma, an initial exposure to allergen results in T(H)2 cell-dependent stimulation of the immune response that mediates the production of IgE and cytokines. Re-exposure to allergen then activates mast cells, which release mediators such as histamines and leukotrienes that recruit other cells, including T(H)2 cells, which mediate the inflammatory response in the lungs. In this minireview, we discuss the current understanding of how associated genetic and environmental factors increase the complexity of allergic asthma and the challenges allergic asthma poses for the development of novel approaches to effective treatment and prevention.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

