Proteomic biomarkers for acute interstitial lung disease in gefitinib-treated Japanese lung cancer patients
- PMID: 21799770
- PMCID: PMC3140475
- DOI: 10.1371/journal.pone.0022062
Proteomic biomarkers for acute interstitial lung disease in gefitinib-treated Japanese lung cancer patients
Abstract
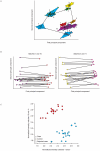
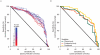
Interstitial lung disease (ILD) events have been reported in Japanese non-small-cell lung cancer (NSCLC) patients receiving EGFR tyrosine kinase inhibitors. We investigated proteomic biomarkers for mechanistic insights and improved prediction of ILD. Blood plasma was collected from 43 gefitinib-treated NSCLC patients developing acute ILD (confirmed by blinded diagnostic review) and 123 randomly selected controls in a nested case-control study within a pharmacoepidemiological cohort study in Japan. We generated ∼7 million tandem mass spectrometry (MS/MS) measurements with extensive quality control and validation, producing one of the largest proteomic lung cancer datasets to date, incorporating rigorous study design, phenotype definition, and evaluation of sample processing. After alignment, scaling, and measurement batch adjustment, we identified 41 peptide peaks representing 29 proteins best predicting ILD. Multivariate peptide, protein, and pathway modeling achieved ILD prediction comparable to previously identified clinical variables; combining the two provided some improvement. The acute phase response pathway was strongly represented (17 of 29 proteins, p = 1.0×10(-25)), suggesting a key role with potential utility as a marker for increased risk of acute ILD events. Validation by Western blotting showed correlation for identified proteins, confirming that robust results can be generated from an MS/MS platform implementing strict quality control.
Conflict of interest statement
Figures





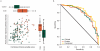
References
-
- American Thoracic Society, European Respiratory Society. American Thoracic Society/European Respiratory Society International Multidisciplinary Consensus Classification of the Idiopathic Interstitial Pneumonias. This joint statement of the American Thoracic Society (ATS), and the European Respiratory Society (ERS) was adopted by the ATS board of directors, June 2001 and by the ERS Executive Committee, June 2001. Am J Respir Crit Care Med. 2002;165:277–304. - PubMed
-
- Inoue A, Saijo Y, Maemondo M, Gomi K, Tokue Y, et al. Severe acute interstitial pneumonia and gefitinib. Lancet. 2003;361:137–139. - PubMed
-
- Kudoh S, Kato H, Nishiwaki Y, Fukuoka M, Nakata K, et al. Interstitial lung disease in Japanese patients with lung cancer: a cohort and nested case-control study. Am J Respir Crit Care Med. 2008;177:1348–1357. - PubMed
-
- Wells AU, Hogaboam CM. Update in diffuse parenchymal lung disease 2006. Am J Respir Crit Care Med. 2007;175:655–660. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

