Using mitochondrial and nuclear sequence data for disentangling population structure in complex pest species: a case study with Dermanyssus gallinae
- PMID: 21799818
- PMCID: PMC3143145
- DOI: 10.1371/journal.pone.0022305
Using mitochondrial and nuclear sequence data for disentangling population structure in complex pest species: a case study with Dermanyssus gallinae
Abstract
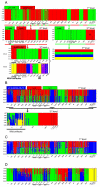
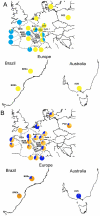
Among global changes induced by human activities, association of breakdown of geographical barriers and impoverishered biodiversity of agroecosystems may have a strong evolutionary impact on pest species. As a consequence of trade networks' expansion, secondary contacts between incipient species, if hybrid incompatibility is not yet reached, may result in hybrid swarms, even more when empty niches are available as usual in crop fields and farms. By providing important sources of genetic novelty for organisms to adapt in changing environments, hybridization may be strongly involved in the emergence of invasive populations. Because national and international trade networks offered multiple hybridization opportunities during the previous and current centuries, population structure of many pest species is expected to be the most intricate and its inference often blurred when using fast-evolving markers. Here we show that mito-nuclear sequence datasets may be the most helpful in disentangling successive layers of admixture in the composition of pest populations. As a model we used D. gallinae s. l., a mesostigmatid mite complex of two species primarily parasitizing birds, namely D. gallinae L1 and D. gallinae s. str. The latter is a pest species, considered invading layer farms in Brazil. The structure of the pest as represented by isolates from both wild and domestic birds, from European (with a focus on France), Australian and Brazilian farms, revealed past hybridization events and very recent contact between deeply divergent lineages. The role of wild birds in the dissemination of mites appears to be null in European and Australian farms, but not in Brazilian ones. In French farms, some recent secondary contact is obviously consecutive to trade flows. Scenarios of populations' history were established, showing five different combinations of more or less dramatic bottlenecks and founder events, nearly interspecific hybridizations and recent population mixing within D. gallinae s. str.
Conflict of interest statement
Figures

References
-
- Mack RN, Simberloff D, Lonsdale WM, Evans H, Clout M, et al. Biotic invasions: causes, epidemiology, global consequences, and control. Ecol appl. 2000;10:689–710.
-
- Huber DM, Hugh-Jones ME, Rust MK, Sheffield SR, Simberloff D, et al. Invasive pest species: impacts on agricultural production, natural resources, and the environment. 2002. Council for Agricultural Science and Technology (CAST) no. 20 ( http://library.wur.nl/journals/local/cast.htm)
-
- Kneitel JM, Perrault D. Disturbance-induced changes in community composition increase species invasion success. Community ecol. 2006;7:245–252.
-
- Lee CE. Evolutionary genetics of invasive species. Trends ecol evol. 2002;17:386–391.
-
- Seehausen O, Takimoto G, Roy D, Jokela J. Speciation reversal and biodiversity dynamics with hybridization in changing environments. Mol ecol. 2008;17:30–44. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

