Genome mining demonstrates the widespread occurrence of gene clusters encoding bacteriocins in cyanobacteria
- PMID: 21799841
- PMCID: PMC3140520
- DOI: 10.1371/journal.pone.0022384
Genome mining demonstrates the widespread occurrence of gene clusters encoding bacteriocins in cyanobacteria
Abstract
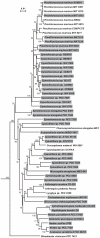
Cyanobacteria are a rich source of natural products with interesting biological activities. Many of these are peptides and the end products of a non-ribosomal pathway. However, several cyanobacterial peptide classes were recently shown to be produced through the proteolytic cleavage and post-translational modification of short precursor peptides. A new class of bacteriocins produced through the proteolytic cleavage and heterocyclization of precursor proteins was recently identified from marine cyanobacteria. Here we show the widespread occurrence of bacteriocin gene clusters in cyanobacteria through comparative analysis of 58 cyanobacterial genomes. A total of 145 bacteriocin gene clusters were discovered through genome mining. These clusters encoded 290 putative bacteriocin precursors. They ranged in length from 28 to 164 amino acids with very little sequence conservation of the core peptide. The gene clusters could be classified into seven groups according to their gene organization and domain composition. This classification is supported by phylogenetic analysis, which further indicated independent evolutionary trajectories of gene clusters in different groups. Our data suggests that cyanobacteria are a prolific source of low-molecular weight post-translationally modified peptides.
Conflict of interest statement
Figures
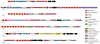



References
-
- Riley MA, Wertz JE. Bacteriocins: Evolution, ecology, and application. Annu Rev Microbiol. 2002;56:117–137. - PubMed
-
- Riley MA, Wertz JE. Bacteriocin diversity: Ecological and evolutionary perspectives. Biochimie. 2002;84:357–364. - PubMed
-
- Ha???varstein LS, Diep DB, Nes IF. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol Microbiol. 1995;16:229–240. - PubMed
-
- Willey JM, van der Donk WA. Lantibiotics: Peptides of diverse structure and function. Annu Rev Microbiol. 2007;61:477–501. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

