Cellular and viral factors regulating Merkel cell polyomavirus replication
- PMID: 21799863
- PMCID: PMC3142164
- DOI: 10.1371/journal.pone.0022468
Cellular and viral factors regulating Merkel cell polyomavirus replication
Abstract
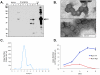
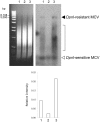
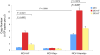
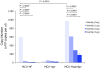
Merkel cell polyomavirus (MCV), a previously unrecognized component of the human viral skin flora, was discovered as a mutated and clonally-integrated virus inserted into Merkel cell carcinoma (MCC) genomes. We reconstructed a replicating MCV clone (MCV-HF), and then mutated viral sites required for replication or interaction with cellular proteins to examine replication efficiency and viral gene expression. Three days after MCV-HF transfection into 293 cells, although replication is not robust, encapsidated viral DNA and protein can be readily isolated by density gradient centrifugation and typical ∼40 nm diameter polyomavirus virions are identified by electron microscopy. The virus has an orderly gene expression cascade during replication in which large T (LT) and 57kT proteins are first expressed by day 2, followed by expression of small T (sT) and VP1 proteins. VP1 and sT proteins are not detected, and spliced 57kT is markedly diminished, in the replication-defective virus suggesting that early gene splicing and late gene transcription may be dependent on viral DNA replication. MCV replication and encapsidation is increased by overexpression of MCV sT, consistent with sT being a limiting factor during virus replication. Mutation of the MCV LT vacuolar sorting protein hVam6p (Vps39) binding site also enhances MCV replication while exogenous hVam6p overexpression reduces MCV virion production by >90%. Although MCV-HF generates encapsidated wild-type MCV virions, we did not find conditions for persistent transmission to recipient cell lines suggesting that MCV has a highly restricted tropism. These studies identify and highlight the role of polyomavirus DNA replication in viral gene expression and show that viral sT and cellular hVam6p are important factors regulating MCV replication. MCV-HF is a molecular clone that can be readily manipulated to investigate factors affecting MCV replication.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

