Identification of RNF213 as a susceptibility gene for moyamoya disease and its possible role in vascular development
- PMID: 21799892
- PMCID: PMC3140517
- DOI: 10.1371/journal.pone.0022542
Identification of RNF213 as a susceptibility gene for moyamoya disease and its possible role in vascular development
Abstract
Background: Moyamoya disease is an idiopathic vascular disorder of intracranial arteries. Its susceptibility locus has been mapped to 17q25.3 in Japanese families, but the susceptibility gene is unknown.
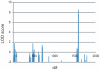
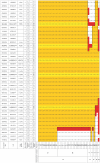
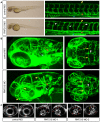
Methodology/principal findings: Genome-wide linkage analysis in eight three-generation families with moyamoya disease revealed linkage to 17q25.3 (P<10(-4)). Fine mapping demonstrated a 1.5-Mb disease locus bounded by D17S1806 and rs2280147. We conducted exome analysis of the eight index cases in these families, with results filtered through Ng criteria. There was a variant of p.N321S in PCMTD1 and p.R4810K in RNF213 in the 1.5-Mb locus of the eight index cases. The p.N321S variant in PCMTD1 could not be confirmed by the Sanger method. Sequencing RNF213 in 42 index cases confirmed p.R4810K and revealed it to be the only unregistered variant. Genotyping 39 SNPs around RNF213 revealed a founder haplotype transmitted in 42 families. Sequencing the 260-kb region covering the founder haplotype in one index case did not show any coding variants except p.R4810K. A case-control study demonstrated strong association of p.R4810K with moyamoya disease in East Asian populations (251 cases and 707 controls) with an odds ratio of 111.8 (P = 10(-119)). Sequencing of RNF213 in East Asian cases revealed additional novel variants: p.D4863N, p.E4950D, p.A5021V, p.D5160E, and p.E5176G. Among Caucasian cases, variants p.N3962D, p.D4013N, p.R4062Q and p.P4608S were identified. RNF213 encodes a 591-kDa cytosolic protein that possesses two functional domains: a Walker motif and a RING finger domain. These exhibit ATPase and ubiquitin ligase activities. Although the mutant alleles (p.R4810K or p.D4013N in the RING domain) did not affect transcription levels or ubiquitination activity, knockdown of RNF213 in zebrafish caused irregular wall formation in trunk arteries and abnormal sprouting vessels.
Conclusions/significance: We provide evidence suggesting, for the first time, the involvement of RNF213 in genetic susceptibility to moyamoya disease.
Conflict of interest statement
Figures






References
-
- Takeuchi K, Shimizu K. Hypogenesis of bilateral internal carotid arteries. Brain Nerve. 1957;9:37–43.
-
- Suzuki J, Takaku A. Cerebrovascular “moyamoya” disease. Disease showing abnormal net-like vessels in base of brain. Arch Neurol. 1969;20:288–299. - PubMed
-
- Goto Y, Yonekawa Y. Worldwide distribution of moyamoya disease. Neurol Med Chir (Tokyo) 1992;32:883–886. - PubMed
-
- Kuroda S, Houkin K. Moyamoya disease: current concepts and future perspectives. Lancet Neurol. 2008;7:1056–1066. - PubMed
-
- Miao W, Zhao PL, Zhang YS, Liu HY, Chang Y, et al. Epidemiological and clinical features of Moyamoya disease in Nanjing, China. Clin Neurol Neurosurg. 2010;112:199–203. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

