Epigenetic silencing of nucleolar rRNA genes in Alzheimer's disease
- PMID: 21799908
- PMCID: PMC3142181
- DOI: 10.1371/journal.pone.0022585
Epigenetic silencing of nucleolar rRNA genes in Alzheimer's disease
Abstract
Background: Ribosomal deficits are documented in mild cognitive impairment (MCI), which often represents an early stage Alzheimer's disease (AD), as well as in advanced AD. The nucleolar rRNA genes (rDNA), transcription of which is critical for ribosomal biogenesis, are regulated by epigenetic silencing including promoter CpG methylation.
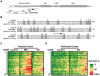
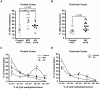
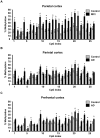
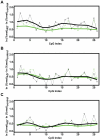
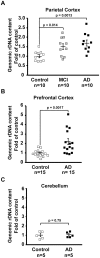
Methodology/principal findings: To assess whether CpG methylation of the rDNA promoter was dysregulated across the AD spectrum, we analyzed brain samples from 10 MCI-, 23 AD-, and, 24 age-matched control individuals using bisulfite mapping. The rDNA promoter became hypermethylated in cerebro-cortical samples from MCI and AD groups. In parietal cortex, the rDNA promoter was hypermethylated more in MCI than in advanced AD. The cytosine methylation of total genomic DNA was similar in AD, MCI, and control samples. Consistent with a notion that hypermethylation-mediated silencing of the nucleolar chromatin stabilizes rDNA loci, preventing their senescence-associated loss, genomic rDNA content was elevated in cerebrocortical samples from MCI and AD groups.
Conclusions/significance: In conclusion, rDNA hypermethylation could be a new epigenetic marker of AD. Moreover, silencing of nucleolar chromatin may occur during early stages of AD pathology and play a role in AD-related ribosomal deficits and, ultimately, dementia.
Conflict of interest statement
Figures
References
-
- Szyf M. Epigenetics, DNA methylation, and chromatin modifying drugs. Annu Rev Pharmacol Toxicol. 2009;49:243–263. - PubMed
-
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. - PubMed
-
- Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. - PubMed
-
- Chouliaras L, Rutten BP, Kenis G, Peerbooms O, Visser PJ, et al. Epigenetic regulation in the pathophysiology of Alzheimer's disease. Prog Neurobiol. 2010;90:498–510. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

