Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology
- PMID: 21799941
- PMCID: PMC3140518
- DOI: 10.1371/journal.pone.0022751
Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology
Abstract
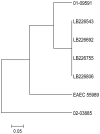
An ongoing outbreak of exceptionally virulent Shiga toxin (Stx)-producing Escherichia coli O104:H4 centered in Germany, has caused over 830 cases of hemolytic uremic syndrome (HUS) and 46 deaths since May 2011. Serotype O104:H4, which has not been detected in animals, has rarely been associated with HUS in the past. To prospectively elucidate the unique characteristics of this strain in the early stages of this outbreak, we applied whole genome sequencing on the Life Technologies Ion Torrent PGM™ sequencer and Optical Mapping to characterize one outbreak isolate (LB226692) and a historic O104:H4 HUS isolate from 2001 (01-09591). Reference guided draft assemblies of both strains were completed with the newly introduced PGM™ within 62 hours. The HUS-associated strains both carried genes typically found in two types of pathogenic E. coli, enteroaggregative E. coli (EAEC) and enterohemorrhagic E. coli (EHEC). Phylogenetic analyses of 1,144 core E. coli genes indicate that the HUS-causing O104:H4 strains and the previously published sequence of the EAEC strain 55989 show a close relationship but are only distantly related to common EHEC serotypes. Though closely related, the outbreak strain differs from the 2001 strain in plasmid content and fimbrial genes. We propose a model in which EAEC 55989 and EHEC O104:H4 strains evolved from a common EHEC O104:H4 progenitor, and suggest that by stepwise gain and loss of chromosomal and plasmid-encoded virulence factors, a highly pathogenic hybrid of EAEC and EHEC emerged as the current outbreak clone. In conclusion, rapid next-generation technologies facilitated prospective whole genome characterization in the early stages of an outbreak.
Conflict of interest statement
Figures




References
-
- Mellmann A, Bielaszewska M, Karch H. Intrahost genome alterations in enterohemorrhagic Escherichia coli. Gastroenterology. 2009;136:1925–1938. - PubMed
-
- Tarr PI, Gordon CA, Chandler WL. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet. 2005;365:1073–1086. - PubMed
-
- Brzuszkiewicz E, Thurmer A, Schuldes J, Leimbach A, Liesegang H, et al. Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: Entero-Aggregative-Haemorrhagic Escherichia coli (EAHEC). 2011. Arch Microbiol 2011 Jun 29. [Epub ahead of print] - PMC - PubMed
-
- Askar M, Faber MS, Frank C, Bernard H, Gilsdorf A, et al. Update on the ongoing outbreak of haemolytic uraemic syndrome due to Shiga toxin-producing Escherichia coli (STEC) serotype O104, Germany, May 2011. Euro Surveill. 2011;16:pii = 19883. - PubMed
-
- Frank C, Werber D, Cramer JP, Askar M, Faber M, et al. Epidemic Profile of Shiga-Toxin-Producing Escherichia coli O104:H4 Outbreak in Germany - Preliminary Report. N Engl J Med. 2011. In press. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

