Genomic approaches to deconstruct pluripotency
- PMID: 21801025
- PMCID: PMC3652340
- DOI: 10.1146/annurev-genom-082410-101506
Genomic approaches to deconstruct pluripotency
Abstract
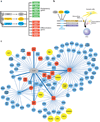
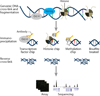
Embryonic stem cells (ESCs) first derived from the inner cell mass of blastocyst-stage embryos have the unique capacity of indefinite self-renewal and potential to differentiate into all somatic cell types. Similar developmental potency can be achieved by reprogramming differentiated somatic cells into induced pluripotent stem cells (iPSCs). Both types of pluripotent stem cells provide great potential for fundamental studies of tissue differentiation, and hold promise for disease modeling, drug development, and regenerative medicine. Although much has been learned about the molecular mechanisms that underlie pluripotency in such cells, our understanding remains incomplete. A comprehensive understanding of ESCs and iPSCs requires the deconstruction of complex transcription regulatory networks, epigenetic mechanisms, and biochemical interactions critical for the maintenance of self-renewal and pluripotency. In this review, we will discuss recent advances gleaned from application of global "omics" techniques to dissect the molecular mechanisms that define the pluripotent state.
Figures
References
-
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, et al. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006;8:532–538. - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

