Evolution of invertebrate deuterostomes and Hox/ParaHox genes
- PMID: 21802045
- PMCID: PMC5054439
- DOI: 10.1016/S1672-0229(11)60011-9
Evolution of invertebrate deuterostomes and Hox/ParaHox genes
Abstract
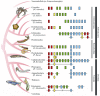
Transcription factors encoded by Antennapedia-class homeobox genes play crucial roles in controlling development of animals, and are often found clustered in animal genomes. The Hox and ParaHox gene clusters have been regarded as evolutionary sisters and evolved from a putative common ancestral gene complex, the ProtoHox cluster, prior to the divergence of the Cnidaria and Bilateria (bilaterally symmetrical animals). The Deuterostomia is a monophyletic group of animals that belongs to the Bilateria, and a sister group to the Protostomia. The deuterostomes include the vertebrates (to which we belong), invertebrate chordates, hemichordates, echinoderms and possibly xenoturbellids, as well as acoelomorphs. The studies of Hox and ParaHox genes provide insights into the origin and subsequent evolution of the bilaterian animals. Recently, it becomes apparent that among the Hox and ParaHox genes, there are significant variations in organization on the chromosome, expression pattern, and function. In this review, focusing on invertebrate deuterostomes, I first summarize recent findings about Hox and ParaHox genes. Next, citing unsolved issues, I try to provide clues that might allow us to reconstruct the common ancestor of deuterostomes, as well as understand the roles of Hox and ParaHox genes in the development and evolution of deuterostomes.
Copyright © 2011 Beijing Genomics Institute. Published by Elsevier Ltd. All rights reserved.
Figures
References
-
- Duboule D. Temporal colinearity and the phylotypic progression: a basis for the stability of a vertebrate Bauplan and the evolution of morphologies through heterochrony. Dev. Suppl. 1994;1994:135–142. - PubMed
-
- Graham A. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell. 1989;57:367–378. - PubMed
-
- Kessel M., Gruss P. Homeotic transformations of murine vertebrae and concomitant alteration of Hox codes induced by retinoic acid. Cell. 1991;67:89–104. - PubMed
-
- Duboule D., Morata G. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. 1994;10:358–364. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

