The roles of transcription and genotoxins underlying p53 mutagenesis in vivo
- PMID: 21803733
- PMCID: PMC3179427
- DOI: 10.1093/carcin/bgr177
The roles of transcription and genotoxins underlying p53 mutagenesis in vivo
Abstract
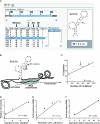
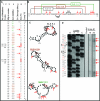
Transcription drives supercoiling which forms and stabilizes single-stranded (ss) DNA secondary structures with loops exposing G and C bases that are intrinsically mutable and vulnerable to non-enzymatic hydrolytic reactions. Since many studies in prokaryotes have shown direct correlations between the frequencies of transcription and mutation, we conducted in silico analyses using the computer program, mfg, which simulates transcription and predicts the location of known mutable bases in loops of high-stability secondary structures. Mfg analyses of the p53 tumor suppressor gene predicted the location of mutable bases and mutation frequencies correlated with the extent to which these mutable bases were exposed in secondary structures. In vitro analyses have now confirmed that the 12 most mutable bases in p53 are in fact located in predicted ssDNA loops of these structures. Data show that genotoxins have two independent effects on mutagenesis and the incidence of cancer: Firstly, they activate p53 transcription, which increases the number of exposed mutable bases and also increases mutation frequency. Secondly, genotoxins increase the frequency of G-to-T transversions resulting in a decrease in G-to-A and C mutations. This precise compensatory shift in the 'fate' of G mutations has no impact on mutation frequency. Moreover, it is consistent with our proposed mechanism of mutagenesis in which the frequency of G exposure in ssDNA via transcription is rate limiting for mutation frequency in vivo.
Figures

References
-
- Hellin A, et al. Nuclear factor—κB-dependent regulation of p53 gene expression induced by daunomycin genotoxic drug. Oncoene. 1998;16:1187–1195. - PubMed
-
- Li H, et al. p53 promoter-based reporter gene in vitro assays for quick assessment of agents with genotoxic potential. Acta Biochim. Biophys. Sin. (Shanghai) 2007;39:181–186. - PubMed
-
- Pe XH, et al. Benzo[a]pyrene activates the human p53 gene through induction of nuclear factor kappaB activity. J. Biol. Chem. 1999;274:35240–35246. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

