Genome-wide in silico identification of new conserved and functional retinoic acid receptor response elements (direct repeats separated by 5 bp)
- PMID: 21803772
- PMCID: PMC3190930
- DOI: 10.1074/jbc.M111.263681
Genome-wide in silico identification of new conserved and functional retinoic acid receptor response elements (direct repeats separated by 5 bp)
Abstract
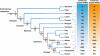
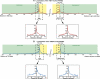
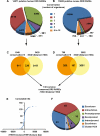
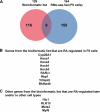
The nuclear retinoic acid receptors interact with specific retinoic acid (RA) response elements (RAREs) located in the promoters of target genes to orchestrate transcriptional networks involved in cell growth and differentiation. Here we describe a genome-wide in silico analysis of consensus DR5 RAREs based on the recurrent RGKTSA motifs. More than 15,000 DR5 RAREs were identified and analyzed for their localization and conservation in vertebrates. We selected 138 elements located ±10 kb from transcription start sites and gene ends and conserved across more than 6 species. We also validated the functionality of these RAREs by analyzing their ability to bind retinoic acid receptors (ChIP sequencing experiments) as well as the RA regulation of the corresponding genes (RNA sequencing and quantitative real time PCR experiments). Such a strategy provided a global set of high confidence RAREs expanding the known experimentally validated RAREs repertoire associated to a series of new genes involved in cell signaling, development, and tumor suppression. Finally, the present work provides a valuable knowledge base for the analysis of a wider range of RA-target genes in different species.
Figures




References
-
- Bour G., Taneja R., Rochette-Egly C. (2006) in Nuclear Receptors in Development (Taneja R. ed.) pp. 211–253, Elsevier Science Publishing Co., Inc., New York
-
- Duong V., Rochette-Egly C. (2011) Biochim. Biophys. Acta 1812, 1023–1031 - PubMed
-
- Samarut E., Rochette-Egly C. (2011) Mol. Cell. Endocrinol., in press - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

