Stem-loop structures can effectively substitute for an RNA pseudoknot in -1 ribosomal frameshifting
- PMID: 21803791
- PMCID: PMC3203594
- DOI: 10.1093/nar/gkr579
Stem-loop structures can effectively substitute for an RNA pseudoknot in -1 ribosomal frameshifting
Abstract
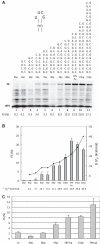
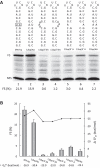
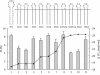
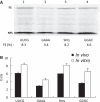
-1 Programmed ribosomal frameshifting (PRF) in synthesizing the gag-pro precursor polyprotein of Simian retrovirus type-1 (SRV-1) is stimulated by a classical H-type pseudoknot which forms an extended triple helix involving base-base and base-sugar interactions between loop and stem nucleotides. Recently, we showed that mutation of bases involved in triple helix formation affected frameshifting, again emphasizing the role of the triple helix in -1 PRF. Here, we investigated the efficiency of hairpins of similar base pair composition as the SRV-1 gag-pro pseudoknot. Although not capable of triple helix formation they proved worthy stimulators of frameshifting. Subsequent investigation of ∼30 different hairpin constructs revealed that next to thermodynamic stability, loop size and composition and stem irregularities can influence frameshifting. Interestingly, hairpins carrying the stable GAAA tetraloop were significantly less shifty than other hairpins, including those with a UUCG motif. The data are discussed in relation to natural shifty hairpins.
Figures

Similar articles
-
Functional analysis of the SRV-1 RNA frameshifting pseudoknot.Nucleic Acids Res. 2010 Nov;38(21):7665-72. doi: 10.1093/nar/gkq629. Epub 2010 Jul 17. Nucleic Acids Res. 2010. PMID: 20639537 Free PMC article.
-
Tertiary Base Triple Formation in the SRV-1 Frameshifting Pseudoknot Stabilizes Secondary Structure Components.Biochemistry. 2020 Nov 24;59(46):4429-4438. doi: 10.1021/acs.biochem.0c00611. Epub 2020 Nov 9. Biochemistry. 2020. PMID: 33166472
-
Solution structure of the pseudoknot of SRV-1 RNA, involved in ribosomal frameshifting.J Mol Biol. 2001 Jul 27;310(5):1109-23. doi: 10.1006/jmbi.2001.4823. J Mol Biol. 2001. PMID: 11501999 Free PMC article.
-
A review on architecture of the gag-pol ribosomal frameshifting RNA in human immunodeficiency virus: a variability survey of virus genotypes.J Biomol Struct Dyn. 2017 Jun;35(8):1629-1653. doi: 10.1080/07391102.2016.1194231. Epub 2016 Aug 2. J Biomol Struct Dyn. 2017. PMID: 27485859 Review.
-
Structure, stability and function of RNA pseudoknots involved in stimulating ribosomal frameshifting.J Mol Biol. 2000 Apr 28;298(2):167-85. doi: 10.1006/jmbi.2000.3668. J Mol Biol. 2000. PMID: 10764589 Free PMC article. Review.
Cited by
-
Mechanisms and implications of programmed translational frameshifting.Wiley Interdiscip Rev RNA. 2012 Sep-Oct;3(5):661-73. doi: 10.1002/wrna.1126. Epub 2012 Jun 19. Wiley Interdiscip Rev RNA. 2012. PMID: 22715123 Free PMC article. Review.
-
A general strategy to inhibiting viral -1 frameshifting based on upstream attenuation duplex formation.Nucleic Acids Res. 2016 Jan 8;44(1):256-66. doi: 10.1093/nar/gkv1307. Epub 2015 Nov 26. Nucleic Acids Res. 2016. PMID: 26612863 Free PMC article.
-
Mechanical unfolding kinetics of the SRV-1 gag-pro mRNA pseudoknot: possible implications for -1 ribosomal frameshifting stimulation.Sci Rep. 2016 Dec 21;6:39549. doi: 10.1038/srep39549. Sci Rep. 2016. PMID: 28000744 Free PMC article.
-
Spacer-length dependence of programmed -1 or -2 ribosomal frameshifting on a U6A heptamer supports a role for messenger RNA (mRNA) tension in frameshifting.Nucleic Acids Res. 2012 Sep 1;40(17):8674-89. doi: 10.1093/nar/gks629. Epub 2012 Jun 28. Nucleic Acids Res. 2012. PMID: 22743270 Free PMC article.
-
HIV-1 frameshift efficiency is primarily determined by the stability of base pairs positioned at the mRNA entrance channel of the ribosome.Nucleic Acids Res. 2013 Feb 1;41(3):1901-13. doi: 10.1093/nar/gks1254. Epub 2012 Dec 16. Nucleic Acids Res. 2013. PMID: 23248007 Free PMC article.
References
-
- Farabaugh PJ. Translational frameshifting: implications for the mechanism of translational frame maintenance. Prog. Nucleic Acid Res. Mol. Biol. 2000;64:131–170. - PubMed
-
- Nixon PL, Giedroc DP. Energetics of a strongly pH dependent RNA tertiary structure in a frameshifting pseudoknot. J. Mol. Biol. 2000;296:659–671. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

