Interaction of Mycobacterium tuberculosis elongation factor Tu with GTP is regulated by phosphorylation
- PMID: 21803988
- PMCID: PMC3187401
- DOI: 10.1128/JB.05469-11
Interaction of Mycobacterium tuberculosis elongation factor Tu with GTP is regulated by phosphorylation
Abstract
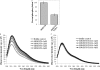
During protein synthesis, translation elongation factor Tu (Ef-Tu) is responsible for the selection and binding of the cognate aminoacyl-tRNA to the acceptor site on the ribosome. The activity of Ef-Tu is dependent on its interaction with GTP. Posttranslational modifications, such as phosphorylation, are known to regulate the activity of Ef-Tu in several prokaryotes. Although a study of the Mycobacterium tuberculosis phosphoproteome showed Ef-Tu to be phosphorylated, the role of phosphorylation in the regulation of Ef-Tu has not been studied. In this report, we show that phosphorylation of M. tuberculosis Ef-Tu (MtbEf-Tu) by PknB reduced its interaction with GTP, suggesting a concomitant reduction in the level of protein synthesis. Overexpression of PknB in Mycobacterium smegmatis indeed reduced the level of protein synthesis. MtbEf-Tu was found to be phosphorylated by PknB on multiple sites, including Thr118, which is required for optimal activity of the protein. We found that kirromycin, an Ef-Tu-specific antibiotic, had a significant effect on the nucleotide binding of unphosphorylated MtbEf-Tu but not on the phosphorylated protein. Our results show that the modulation of the MtbEf-Tu-GTP interaction by phosphorylation can have an impact on cellular protein synthesis and growth. These results also suggest that phosphorylation can change the sensitivity of the protein to the specific inhibitors. Thus, the efficacy of an inhibitor can also depend on the posttranslational modification(s) of the target and should be considered during the development of the molecule.
Figures



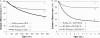
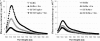
Similar articles
-
Biophysical characterization and ligand-binding properties of the elongation factor Tu from Mycobacterium tuberculosis.Acta Biochim Biophys Sin (Shanghai). 2019 Feb 1;51(2):139-149. doi: 10.1093/abbs/gmy164. Acta Biochim Biophys Sin (Shanghai). 2019. PMID: 30615070
-
Kirromycin drastically reduces the affinity of Escherichia coli elongation factor Tu for aminoacyl-tRNA.Biochemistry. 1991 Jul 9;30(27):6705-10. doi: 10.1021/bi00241a010. Biochemistry. 1991. PMID: 2065055
-
A single amino acid substitution in elongation factor Tu disrupts interaction between the ternary complex and the ribosome.J Bacteriol. 1993 Jan;175(1):240-50. doi: 10.1128/jb.175.1.240-250.1993. J Bacteriol. 1993. PMID: 8416899 Free PMC article.
-
Elongation factor Tu, a GTPase triggered by codon recognition on the ribosome: mechanism and GTP consumption.Biochem Cell Biol. 1995 Nov-Dec;73(11-12):1221-7. doi: 10.1139/o95-132. Biochem Cell Biol. 1995. PMID: 8722040 Review.
-
Structural features in aminoacyl-tRNAs required for recognition by elongation factor Tu.FEBS Lett. 1987 Jun 15;217(2):203-11. doi: 10.1016/0014-5793(87)80664-6. FEBS Lett. 1987. PMID: 3297780 Review.
Cited by
-
Mycobacterium tuberculosis Serine/Threonine Protein Kinases.Microbiol Spectr. 2014 Oct;2(5):10.1128/microbiolspec.MGM2-0006-2013. doi: 10.1128/microbiolspec.MGM2-0006-2013. Microbiol Spectr. 2014. PMID: 25429354 Free PMC article. Review.
-
Phosphorylation regulates mycobacterial proteasome.J Microbiol. 2014 Sep;52(9):743-54. doi: 10.1007/s12275-014-4416-2. Epub 2014 Sep 2. J Microbiol. 2014. PMID: 25224505
-
Ser/Thr phosphorylation as a regulatory mechanism in bacteria.Curr Opin Microbiol. 2015 Apr;24:47-52. doi: 10.1016/j.mib.2015.01.005. Epub 2015 Jan 24. Curr Opin Microbiol. 2015. PMID: 25625314 Free PMC article. Review.
-
TUFM-knockdown inhibits the migration and proliferation of gastrointestinal stromal tumor cells.Oncol Lett. 2020 Nov;20(5):250. doi: 10.3892/ol.2020.12113. Epub 2020 Sep 17. Oncol Lett. 2020. PMID: 32994813 Free PMC article.
-
Genome Wide Phosphoproteome Analysis of Zymomonas mobilis Under Anaerobic, Aerobic, and N2-Fixing Conditions.Front Microbiol. 2019 Sep 4;10:1986. doi: 10.3389/fmicb.2019.01986. eCollection 2019. Front Microbiol. 2019. PMID: 31551951 Free PMC article.
References
-
- Abel K., Jurnak F. 1996. A complex profile of protein elongation: translating chemical energy into molecular movement. Structure 4:229–238 - PubMed
-
- Alexander C., et al. 1995. Phosphorylation of elongation factor Tu prevents ternary complex formation. J. Biol. Chem. 270:14541–14547 - PubMed
-
- Ames G. F., Niakido K. 1979. In vivo methylation of prokaryotic elongation factor Tu. J. Biol. Chem. 254:9947–9950 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

