Choline uptake in Agrobacterium tumefaciens by the high-affinity ChoXWV transporter
- PMID: 21803998
- PMCID: PMC3187443
- DOI: 10.1128/JB.05421-11
Choline uptake in Agrobacterium tumefaciens by the high-affinity ChoXWV transporter
Abstract
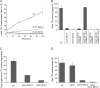
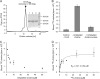
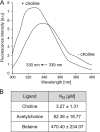
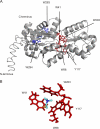
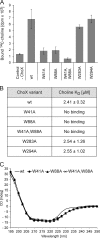
Agrobacterium tumefaciens is a facultative phytopathogen that causes crown gall disease. For successful plant transformation A. tumefaciens requires the membrane lipid phosphatidylcholine (PC), which is produced via the methylation and the PC synthase (Pcs) pathways. The latter route is dependent on choline. Although choline uptake has been demonstrated in A. tumefaciens, the responsible transporter(s) remained elusive. In this study, we identified the first choline transport system in A. tumefaciens. The ABC-type choline transporter is encoded by the chromosomally located choXWV operon (ChoX, binding protein; ChoW, permease; and ChoV, ATPase). The Cho system is not critical for growth and PC synthesis. However, [14C]choline uptake is severely reduced in A. tumefaciens choX mutants. Recombinant ChoX is able to bind choline with high affinity (equilibrium dissociation constant [KD] of ≈2 μM). Since other quaternary amines are bound by ChoX with much lower affinities (acetylcholine, KD of ≈80 μM; betaine, KD of ≈470 μM), the ChoXWV system functions as a high-affinity transporter with a preference for choline. Two tryptophan residues (W40 and W87) located in the predicted ligand-binding pocket are essential for choline binding. The structural model of ChoX built on Sinorhizobium meliloti ChoX resembles the typical structure of substrate binding proteins with a so-called "Venus flytrap mechanism" of substrate binding.
Figures



References
-
- Aktas M., et al. 2010. Phosphatidylcholine biosynthesis and its significance in bacteria interacting with eukaryotic cells. Eur. J. Cell Biol. 89:888–894 - PubMed
-
- Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917 - PubMed
-
- Blusztajn J. K. 1998. Choline, a vital amine. Science 281:794–795 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

