Biodistribution of small interfering RNA at the organ and cellular levels after lipid nanoparticle-mediated delivery
- PMID: 21804077
- PMCID: PMC3261601
- DOI: 10.1369/0022155411410885
Biodistribution of small interfering RNA at the organ and cellular levels after lipid nanoparticle-mediated delivery
Abstract
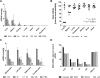
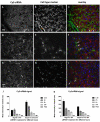
Chemically stabilized small interfering RNA (siRNA) can be delivered systemically by intravenous injection of lipid nanoparticles (LNPs) in rodents and primates. The biodistribution and kinetics of LNP-siRNA delivery in mice at organ and cellular resolution have been studied using immunofluorescence (IF) staining and quantitative polymerase chain reaction (qPCR). At 0.5 and 2 hr post tail vein injection of Cy5-labeled siRNA encapsulated in LNP, the organ rank-order of siRNA levels is liver > spleen > kidney, with only negligible accumulation in duodenum, lung, heart, and brain. Similar conclusions were drawn by using qPCR to measure tissue siRNA levels as a secondary end point. siRNA levels in these tissues decreased by more than 10-fold after 24 hr. Within the liver, LNPs delivered siRNA to hepatocytes, Kupffer cells, and sinusoids in a time-dependent manner, as revealed by IF staining and signal quantitation methods established using OPERA/Columbus software. siRNA first accumulated in liver sinusoids and trafficked to hepatocytes by 2 hr post dose, corresponding to the onset of target mRNA silencing. Fluorescence in situ hybridization methods were used to detect both strands of siRNA in fixed tissues. Collectively, the authors have implemented a platform to evaluate biodistribution of siRNA across cell types and across tissues in vivo, with the objective of elucidating the pharmacokinetic and pharmacodynamic relationship to guide optimization of delivery vehicles.
© The Author(s) 2011
Conflict of interest statement
All authors are full-time employees of Merck & Co, Inc.
Figures



References
-
- Baldwin L, Flanagan BF, Hunt JA. 2005. Flow cytometric measurement of phagocytosis reveals a role for C3b in metal particle uptake by phagocytes. J Biomed Mater Res A. 73:80–85 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

